10k A375 Cells Transduced with (1) Non-Target and (1) Target sgRNA¶
Dataset: 10k A375 Cells Transduced with (1) Non-Target and (1) Target sgRNA, Dual Indexed
The detailed description of this dataset can be found here.
Preparation¶
Download fastq files.
$ wget https://cg.10xgenomics.com/samples/cell-exp/4.0.0/SC3_v3_NextGem_DI_CRISPR_10K/SC3_v3_NextGem_DI_CRISPR_10K_fastqs.tar
$ tar xvf SC3_v3_NextGem_DI_PBMC_CSP_1K/SC3_v3_NextGem_DI_PBMC_CSP_1K_fastqs.tar
Combine reads of different lanes.
$ cat SC3_v3_NextGem_DI_CRISPR_10K_fastqs/SC3_v3_NextGem_DI_CRISPR_10K_crispr_fastqs/SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_L00?_R1_001.fastq.gz > SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_combined_R1_001.fastq.gz
$ cat SC3_v3_NextGem_DI_CRISPR_10K_fastqs/SC3_v3_NextGem_DI_CRISPR_10K_crispr_fastqs/SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_L00?_R2_001.fastq.gz > SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_combined_R2_001.fastq.gz
Download cell barcode info. These are the cell-associated barcodes in this single cell RNA-Seq library.
$ wget https://cf.10xgenomics.com/samples/cell-exp/4.0.0/SC3_v3_NextGem_DI_CRISPR_10K/SC3_v3_NextGem_DI_CRISPR_10K_filtered_feature_bc_matrix.tar.gz
$ tar zxvf SC3_v3_NextGem_DI_CRISPR_10K_filtered_feature_bc_matrix.tar.gz
Inspect cell barcodes.
$ gzip -dc filtered_feature_bc_matrix/barcodes.tsv.gz | head
AAACCCACACATAGCT-1
AAACCCACATCATGAC-1
AAACCCAGTCATCGGC-1
AAACCCAGTCGCACAC-1
AAACCCAGTGCAACGA-1
AAACCCATCAAGCGTT-1
AAACCCATCAGATGCT-1
AAACCCATCATCTACT-1
AAACCCATCCTTATGT-1
AAACCCATCTCGGCTT-1
Prepare feature barcodes.
$ wget https://cf.10xgenomics.com/samples/cell-exp/4.0.0/SC3_v3_NextGem_DI_CRISPR_10K/SC3_v3_NextGem_DI_CRISPR_10K_feature_ref.csv
Inspect feature barcode info.
$ cat SC3_v3_NextGem_DI_CRISPR_10K_feature_ref.csv
id,name,read,pattern,sequence,feature_type,target_gene_id,target_gene_name
RAB1A-2,RAB1A-2,R2,(BC)GTTTAAGAGCTAAGCTGGAA,GCCGGCGAACCAGGAAATA,CRISPR Guide Capture,ENSG00000138069,RAB1A
NON_TARGET-1,NON_TARGET-1,R2,(BC)GTTTAAGAGCTAAGCTGGAA,AACGTGCTGACGATGCGGGC,CRISPR Guide Capture,Non-Targeting,Non-Targeting
Clean up.
$ cut -d ',' -f1,5 SC3_v3_NextGem_DI_CRISPR_10K_feature_ref.csv | sed 's/,/\t/g' | tail -2 > SC3_v3_NextGem_DI_CRISPR_10K_feature_ref.tsv
$ cat SC3_v3_NextGem_DI_CRISPR_10K_feature_ref.tsv
RAB1A-2 GCCGGCGAACCAGGAAATA
NON_TARGET-1 AACGTGCTGACGATGCGGGC
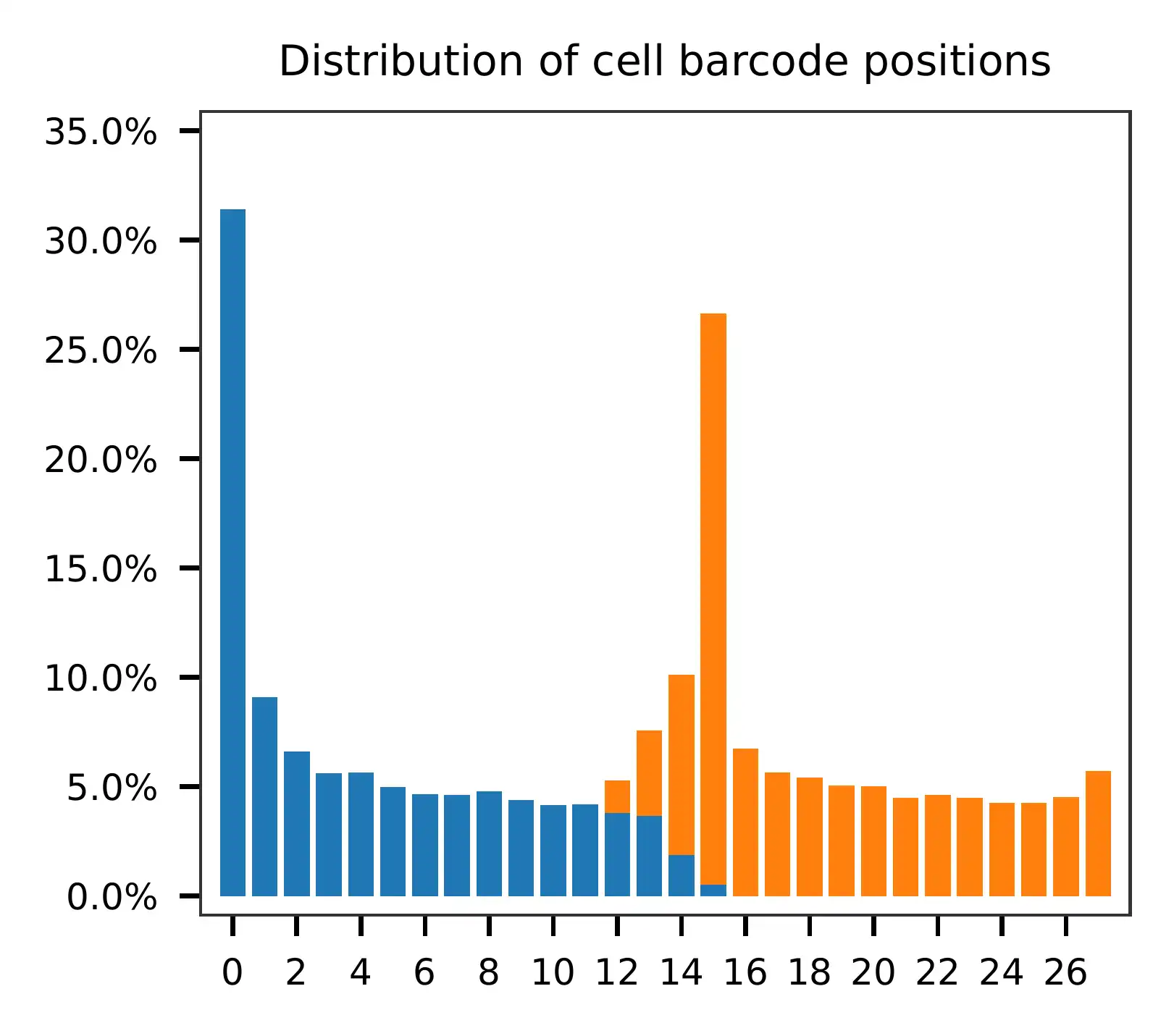
QC¶
The first 20,000 read pairs are sampled (set by -n, default
100,000) for quality control. The -t option can be used to set
the number of threads. By default, diagnostic results and plots are
generated in the qc directory (set by --output_directory), and
the full length of read 1 and read 2 are searched against reference cell
and feature barcodes, respectively. The per base content of both read
pairs and the distribution of matched barcode positions are summarized.
Use -r1_c and/or -r2_c to limit the search range, and -cb_n
and/or -fb_n to set the mismatch tolerance for cell and/or feature
barcode matching (default 3).
$ fba qc \
-1 SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_combined_R1_001.fastq.gz \
-2 SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_combined_R2_001.fastq.gz \
-w filtered_feature_bc_matrix/barcodes.tsv.gz \
-f SC3_v3_NextGem_DI_CRISPR_10K_feature_ref.tsv \
-r1_c 0,16 \
-n 20000
This library was constructed using the Chromium Next GEM Single Cell 3ʹ Reagent Kits v3.1 (Dual Index) with Feature Barcode technology for CRISPR Screening and sequenced on an Illumina NovaSeq 6000. The first 16 bases of read 1 represent cell barcodes, and the following 12 bases represent UMIs. The base content plot indicates that the GC content of cell barcodes is evenly distributed. However, there is a slight T-enrichment in the UMIs.
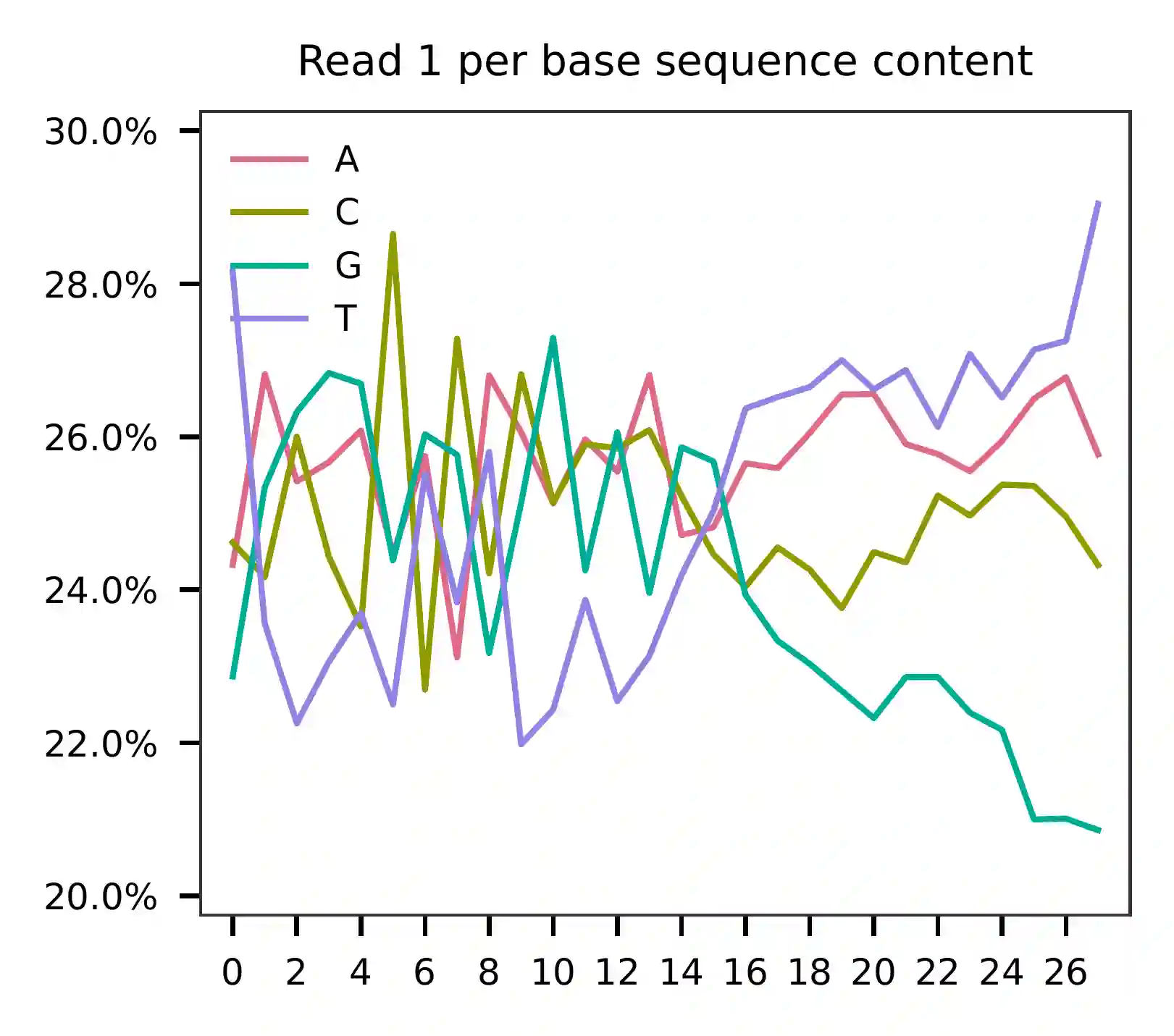
Regarding read 2, the per base content analysis indicates that the first 31 bases are consistent and easily readable. These bases correspond to the Template Switch Oligo (TSO) sequence used in library construction. From base 32 onward, we have observed two distinct genotypes in the sampled reads.
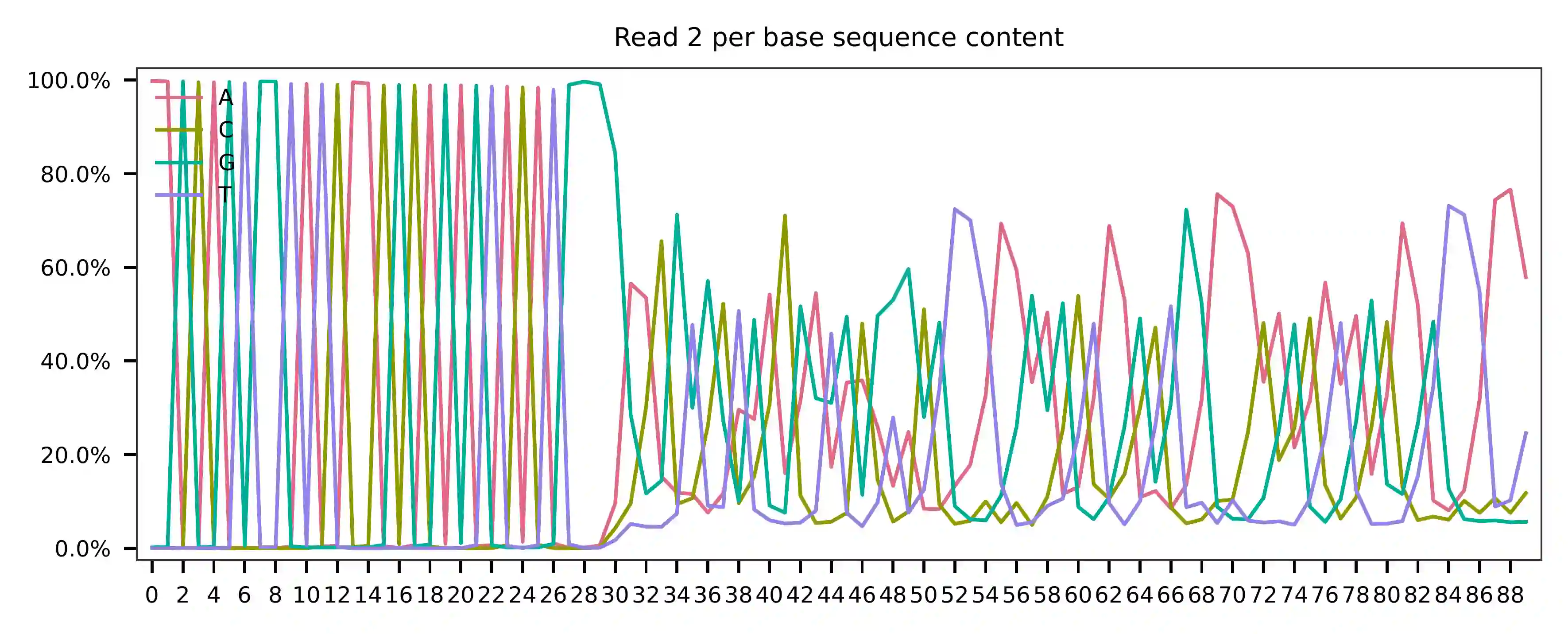
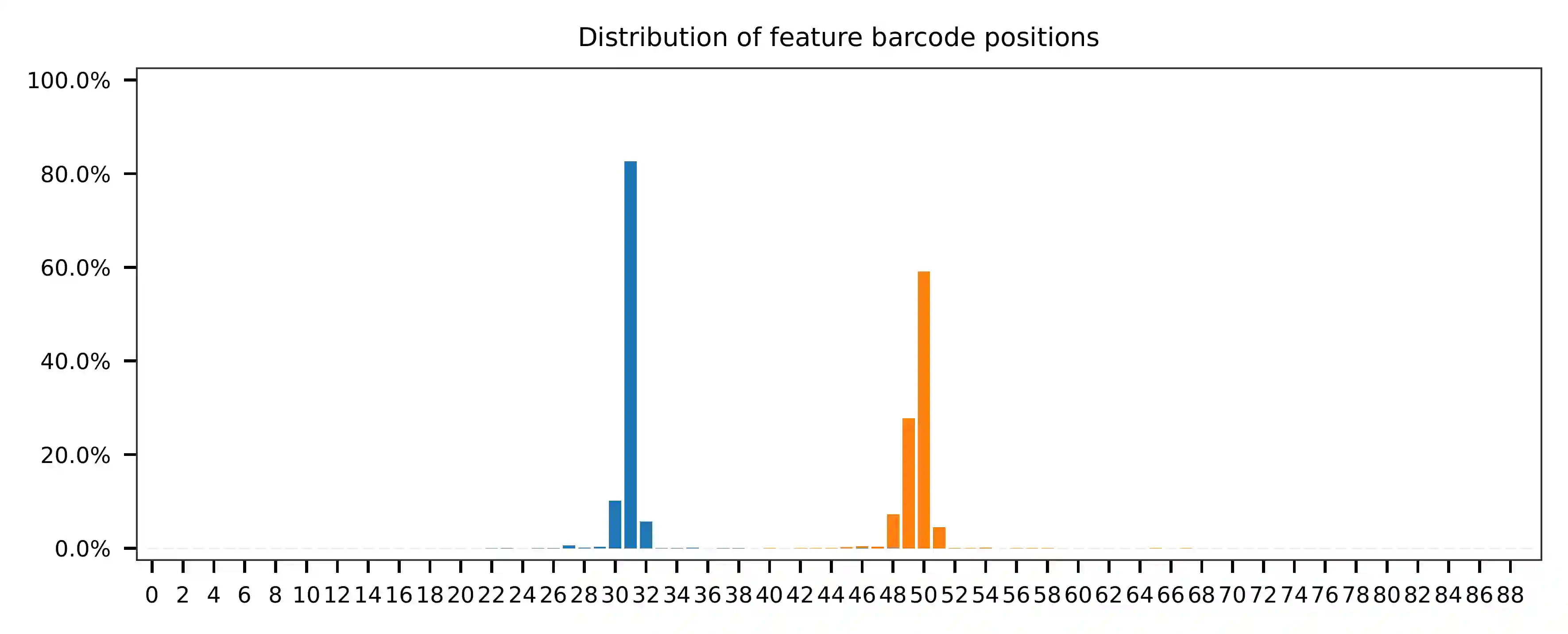
The detailed qc results are stored in the
feature_barcoding_output.tsv.gz file. The matching_pos columns
indicate the matched positions on reads, while the
matching_description columns indicate mismatches in the format of
substitutions:insertions:deletions.
$ gzip -dc qc/feature_barcoding_output.tsv.gz | head
read1_seq cell_barcode cb_matching_pos cb_matching_description read2_seq feature_barcode fb_matching_pos fb_matching_description
CNCCACACACGTGTTAatgagtactagc CCTCACACACGTAGTT 0:15 2:0:1 AAGCAGTGGTATCAACGCAGAGTACATGGGATAGGTTTGGTCCTAGCCTTTCTATTAGCTCTTAGTAAGATTACACATGCAAGCATCCCC no_match NA NA
GNCGCGATCAGCATTActtttgtcaccc GTCGCGAAGAGCATTA 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGACTGTTGCTGGTGTGTACTTGCTAAGGTTTATGTCAGTTCAAGATTATAAGCCCCCCAG no_match NA NA
TNGGAAGGTAAGTGTAatcgagggaaca TGGGAAGCAAAGTGTA 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGGCCGGCGAACCAGGAAATAGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAAT RAB1A-2_GCCGGCGAACCAGGAAATAG 31:51 0:0:0
CNCCCAAGTCGATAGGgagcgcaagcat CCCAACTCACGATAGG 2:16 1:0:2 AAGCAGTGGTATCAACGCAGAGTACATGGGGGCCGGCGAACCAGGAAATAGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAAT RAB1A-2_GCCGGCGAACCAGGAAATAG 31:51 0:0:0
CNCACTGCAAACGGTGggcgtaaatgag CTCACTGGTAACGGTG 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGGCCGGCGAACCAGGAAATAGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAAT RAB1A-2_GCCGGCGAACCAGGAAATAG 31:51 0:0:0
ANCATCACAGGCGCTTgtcccactatat AGCATCAGTGGCGCTT 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGGCCGGCGAACCAGGAAATAGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAAT RAB1A-2_GCCGGCGAACCAGGAAATAG 31:51 0:0:0
ANACGAACACTTTCATccaaaagaagtt AAACGAAGTCTTTCAT 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGGCCGGCGAACCAGGAAATAGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAAT RAB1A-2_GCCGGCGAACCAGGAAATAG 31:51 0:0:0
ANCAACCAGTATCGTTgaaatcctggta AACAACCTCTATCGTT 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGAACGTGCTGACGATGCGGGCGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAA NON_TARGET-1_AACGTGCTGACGATGCGGGC 31:51 0:0:0
GNAGCCCGTACCACATgggcccagtatg GAAGCCCCAACCACAT 0:16 3:0:0 AAGCAGTGGTATCAACGCAGAGTACATGGGGGCCGGCGAACCAGGAAATAGTTTAAGAGCTAAGCTGGAAACAGCATAGCAAGTTTAAAT RAB1A-2_GCCGGCGAACCAGGAAATAG 31:51 0:0:0
Barcode extraction¶
Although the length of the RAB1A-1 and RAB1A-2 feature barcodes differ by one base, they both start at the same position on read 2. To enable accurate feature barcode identification, we will include an extra downstream base (G) for the RAB1A-2 feature barcode to make their lengths equal.
$ cat SC3_v3_NextGem_DI_CRISPR_10K_feature_ref_edited.tsv
RAB1A-2 GCCGGCGAACCAGGAAATAG
NON_TARGET-1 AACGTGCTGACGATGCGGGC
The search ranges for barcode matching are set to 0,16 on read 1 and
31,51 on read 2. We allow for two mismatches for both cell and
feature barcodes using the parameters -cb_m and -cf_m.
$ fba extract \
-1 SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_combined_R1_001.fastq.gz \
-2 SC3_v3_NextGem_DI_CRISPR_10K_crispr_S1_combined_R2_001.fastq.gz \
-w filtered_feature_bc_matrix/barcodes.tsv.gz \
-f SC3_v3_NextGem_DI_CRISPR_10K_feature_ref_edited.tsv \
-o feature_barcoding_output.tsv.gz \
-r1_c 0,16 \
-r2_c 31,51 \
-cb_m 2 \
-fb_m 2
Preview of result.
$ gzip -dc feature_barcoding_output.tsv.gz | head
read1_seq cell_barcode cb_num_mismatches read2_seq feature_barcode fb_num_mismatches
GGCAGTCTCCGTTACTtatccagccttc GGCAGTCTCGGTAACT 2 aagcagtggtatcaacgcagagtacatggggGCCGGCGAACCAGGAAATAGtttaagagctaagctggaaacagcatagcaagtttaaat RAB1A-2_GCCGGCGAACCAGGAAATAG 0
TTACGTTGTGAATCGGgtggggctcttc TTACGTTCAGAATCGG 2 aagcagtggtatcaacgcagagtacatggggAACGTGCTGACGATGCGGGCgtttaagagctaagctggaaacagcatagcaagtttaaa NON_TARGET-1_AACGTGCTGACGATGCGGGC 0
TCGGGCAAGGATTGGTttctactcggaa TCGGGCATCGATTGGT 2 aagcagtggtatcaacgcagagtacatgggaACGTGCTGACGATGCGGGCGtttaagagctaagctggaaacagcatagcaagtttaaat NON_TARGET-1_AACGTGCTGACGATGCGGGC 2
ACAACCACACATCTAGcggcatcatact ACAACCAGTCATCTAG 2 aagcagtggtatcaacgcagagtacatggggCCGGCGAACCAGGAAATAGTttaagagctaagctggaaacagcatagcaagtttaaata RAB1A-2_GCCGGCGAACCAGGAAATAG 2
AGACTCAAGTGCTAGAacagaactggtg AGACTCATCTGCTAGA 2 aagcagtggtatcaacgcagagtacatggggAACGTGCTGACGATGCGGGCgtttaagagctaagctggaaacagcatagcaagtttaaa NON_TARGET-1_AACGTGCTGACGATGCGGGC 0
GAGTTGTTCGAACATTctgcccgacgtc GAGTTGTAGGAACATT 2 aagcagtggtatcaacgcagagtacatggggAACGTGCTGACGATGCGGGCgtttaagagctaagctggaaacagcatagcaagtttaaa NON_TARGET-1_AACGTGCTGACGATGCGGGC 0
AGACTCAGTGGCACAAtgtcagaattca AGACTCACAGGCACAA 2 aagcagtggtatcaacgcagagtacatggggGCCGGCGAACCAGGAAATAGtttaagagctaagctggaaacagcatagcaagtttaaat RAB1A-2_GCCGGCGAACCAGGAAATAG 0
TGCACGGAGGATAACCcgtgcacgtaca TGCACGGTCGATAACC 2 aagcagtggtatcaacgcagagtacatggggGCCGGCGAACCAGGAAATAGtttaagagctaagctggaaacagcatagcaagtttaaat RAB1A-2_GCCGGCGAACCAGGAAATAG 0
CGTAGTAGTAACACGGaagagggaactg CGTAGTAGTAACGCGA 2 aagcagtggtatcaacgcagagtacatggggAACGTGCTGACGATGCGGGCgtttaagagctaagctggaaacagcatagcaagtttaaa NON_TARGET-1_AACGTGCTGACGATGCGGGC 0
Result summary.
64.7% (93,795,979 out of 145,032,428) of total read pairs have valid cell and feature barcodes. Majority of fragments in this library have correct structure.
2021-02-15 01:51:59,262 - fba.__main__ - INFO - fba version: 0.0.7
2021-02-15 01:51:59,262 - fba.__main__ - INFO - Initiating logging ...
2021-02-15 01:51:59,262 - fba.__main__ - INFO - Python version: 3.7
2021-02-15 01:51:59,262 - fba.__main__ - INFO - Using extract subcommand ...
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Number of reference cell barcodes: 11,791
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Number of reference feature barcodes: 2
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Read 1 coordinates to search: [0, 16)
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Read 2 coordinates to search: [31, 51)
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Cell barcode maximum number of mismatches: 2
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Feature barcode maximum number of mismatches: 2
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Read 1 maximum number of N allowed: 3
2021-02-15 01:51:59,276 - fba.levenshtein - INFO - Read 2 maximum number of N allowed: 3
2021-02-15 01:52:02,510 - fba.levenshtein - INFO - Matching ...
2021-02-15 02:20:39,807 - fba.levenshtein - INFO - Read pairs processed: 10,000,000
2021-02-15 02:49:04,142 - fba.levenshtein - INFO - Read pairs processed: 20,000,000
2021-02-15 03:17:27,422 - fba.levenshtein - INFO - Read pairs processed: 30,000,000
2021-02-15 03:45:54,615 - fba.levenshtein - INFO - Read pairs processed: 40,000,000
2021-02-15 04:14:23,049 - fba.levenshtein - INFO - Read pairs processed: 50,000,000
2021-02-15 04:42:49,377 - fba.levenshtein - INFO - Read pairs processed: 60,000,000
2021-02-15 05:11:15,736 - fba.levenshtein - INFO - Read pairs processed: 70,000,000
2021-02-15 05:39:43,011 - fba.levenshtein - INFO - Read pairs processed: 80,000,000
2021-02-15 06:08:09,940 - fba.levenshtein - INFO - Read pairs processed: 90,000,000
2021-02-15 06:36:39,658 - fba.levenshtein - INFO - Read pairs processed: 100,000,000
2021-02-15 07:05:08,115 - fba.levenshtein - INFO - Read pairs processed: 110,000,000
2021-02-15 07:33:32,101 - fba.levenshtein - INFO - Read pairs processed: 120,000,000
2021-02-15 08:02:01,233 - fba.levenshtein - INFO - Read pairs processed: 130,000,000
2021-02-15 08:30:29,660 - fba.levenshtein - INFO - Read pairs processed: 140,000,000
2021-02-15 08:44:47,038 - fba.levenshtein - INFO - Number of read pairs processed: 145,032,428
2021-02-15 08:44:47,038 - fba.levenshtein - INFO - Number of read pairs w/ valid barcodes: 93,795,979
2021-02-15 08:44:47,153 - fba.__main__ - INFO - Done.
Matrix generation¶
Only fragments with correctly matched cell and feature barcodes are
included, while fragments with UMI lengths less than the specified value
are discarded. UMI removal is performed using UMI-tools (Smith, T., et
al. 2017. Genome Res. 27, 491–499.), with the starting position on
read 1 set by -us (default 16) and the length set by -ul
(default 12). The UMI deduplication method can be set using -ud
(default directional), and the UMI deduplication mismatch threshold
can be specified using -um (default 1).
The generated feature count matrix can be easily imported into well-established single cell analysis packages: Seurat and Scanpy.
$ fba count \
-i feature_barcoding_output.tsv.gz \
-o matrix_featurecount.csv.gz \
-us 16 \
-ul 12 \
-um 1 \
-ud directional
Result summary.
7.6% (7,145,799 out of 93,795,979) of read pairs with valid cell and feature barcodes are unique fragments. 4.9% (7,143,943 out of 145,032,428) of total sequenced read pairs contribute to the final matrix.
2020-10-20 04:47:32,738 - fba.__main__ - INFO - fba version: 0.0.7
2020-10-20 04:47:32,738 - fba.__main__ - INFO - Initiating logging ...
2020-10-20 04:47:32,738 - fba.__main__ - INFO - Python version: 3.7
2020-10-20 04:47:32,738 - fba.__main__ - INFO - Using count subcommand ...
2020-10-20 04:47:32,738 - fba.count - INFO - UMI-tools version: 1.0.1
2020-10-20 04:47:32,795 - fba.count - INFO - UMI starting position on read 1: 16
2020-10-20 04:47:32,795 - fba.count - INFO - UMI length: 12
2020-10-20 04:47:32,795 - fba.count - INFO - UMI-tools deduplication threshold: 1
2020-10-20 04:47:32,795 - fba.count - INFO - UMI-tools deduplication method: directional
2020-10-20 04:47:32,795 - fba.count - INFO - Header line: read1_seq cell_barcode cb_num_mismatches read2_seq feature_barcode fb_num_mismatches
2020-10-20 04:51:50,886 - fba.count - INFO - Number of lines processed: 93,795,979
2020-10-20 04:51:50,893 - fba.count - INFO - Number of cell barcodes detected: 11,758
2020-10-20 04:51:50,894 - fba.count - INFO - Number of features detected: 2
2020-10-20 05:00:42,298 - fba.count - INFO - Total UMIs after deduplication: 7,145,799
2020-10-20 05:00:42,320 - fba.count - INFO - Median number of UMIs per cell: 477.0
2020-10-20 05:00:42,434 - fba.__main__ - INFO - Done.
Demultiplexing¶
Negative binomial distribution¶
Cells are demultiplexed based on the feature count matrix using
demultiplexing method 1 (set by -dm), which is implemented based
on the method described by Stoeckius, M., et al. (2018) with some
modifications. The output directory for demultiplexing is set by
--output_directory (default demultiplexed). A cell identity
matrix is generated, where 0 indicates negative and 1 indicates
positive. To adjust the quantile threshold for demultiplexing, use
-q (default 0.9999). To generate visualization plots, set
-v.
$ fba demultiplex \
-i matrix_featurecount.csv.gz \
--output_directory demultiplexed \
-dm 1 \
-q 0.75 \
-v
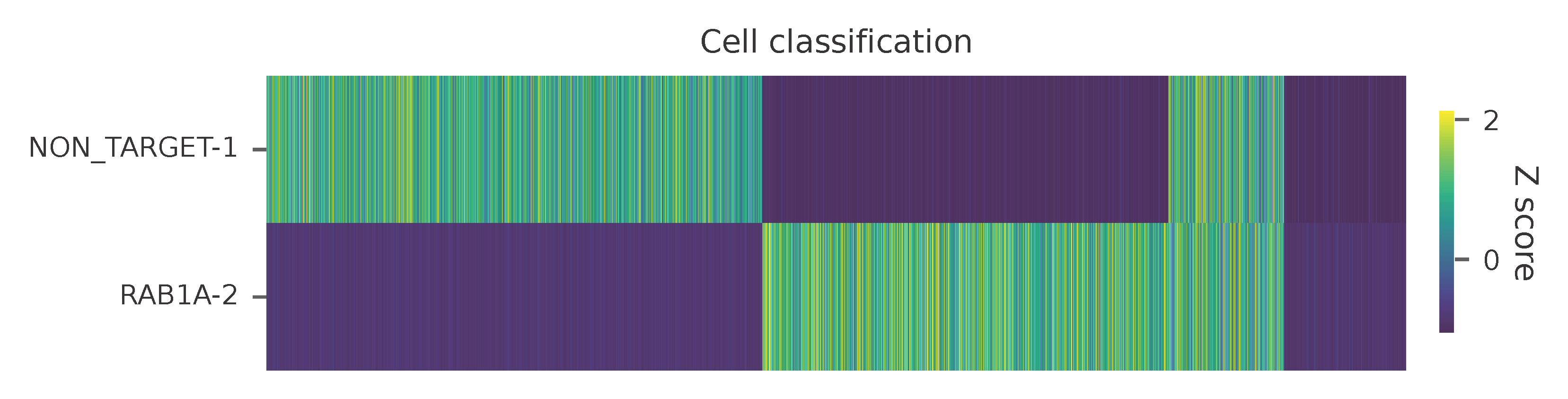
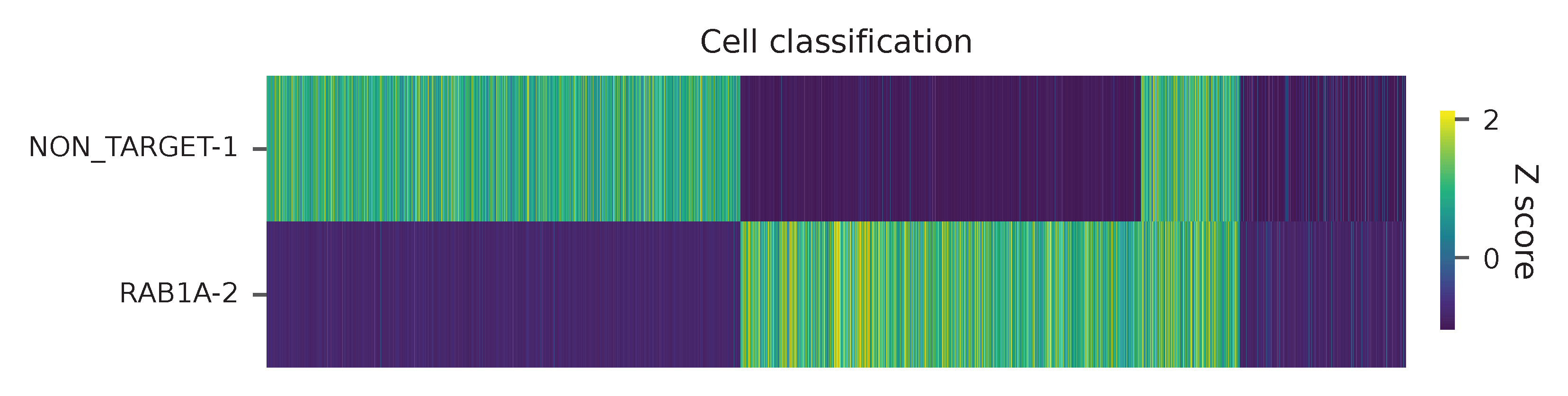
Heatmap of the relative abundance of features (sgRNAs) across all cells. Each column represents a single cell.
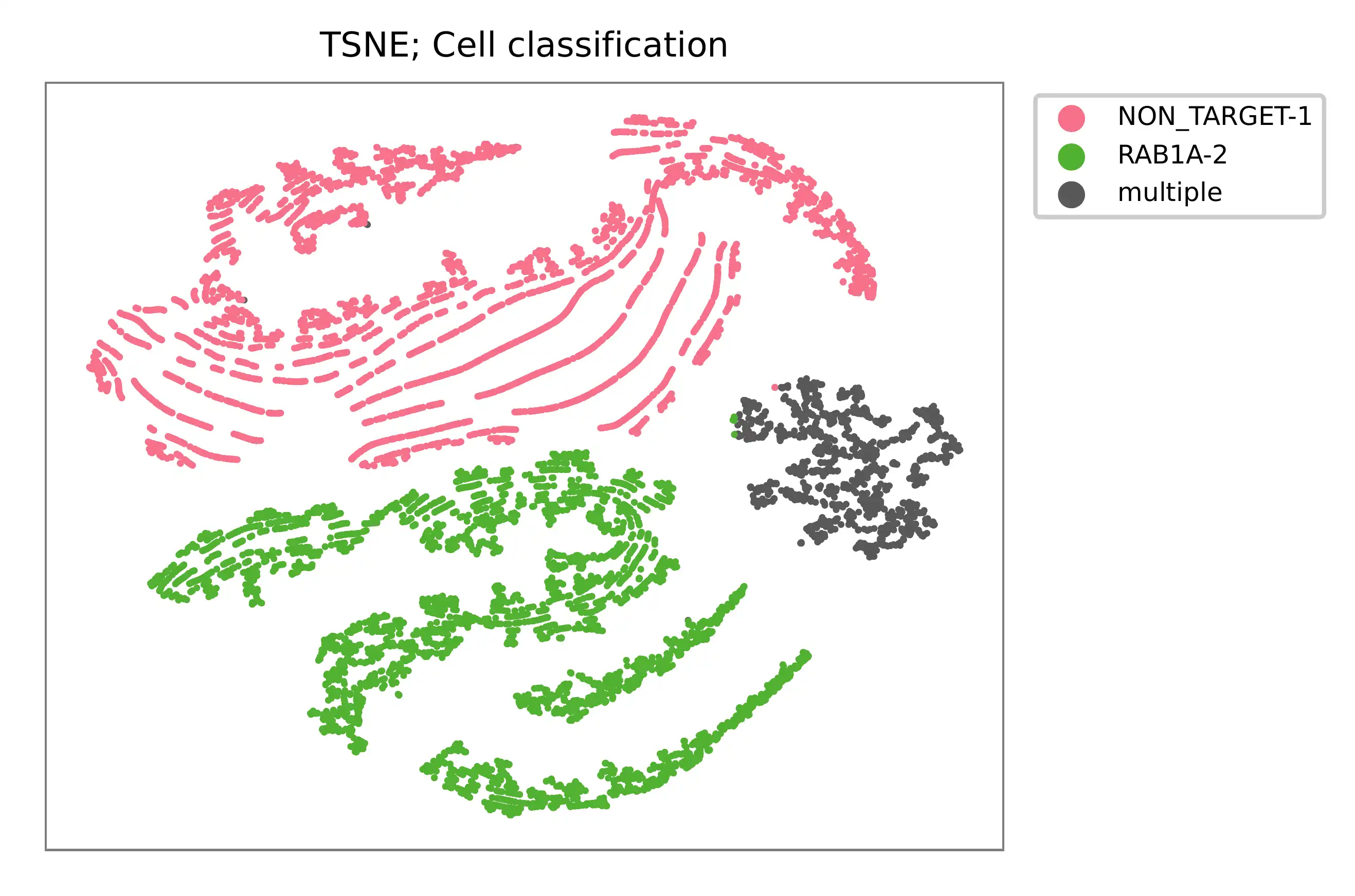
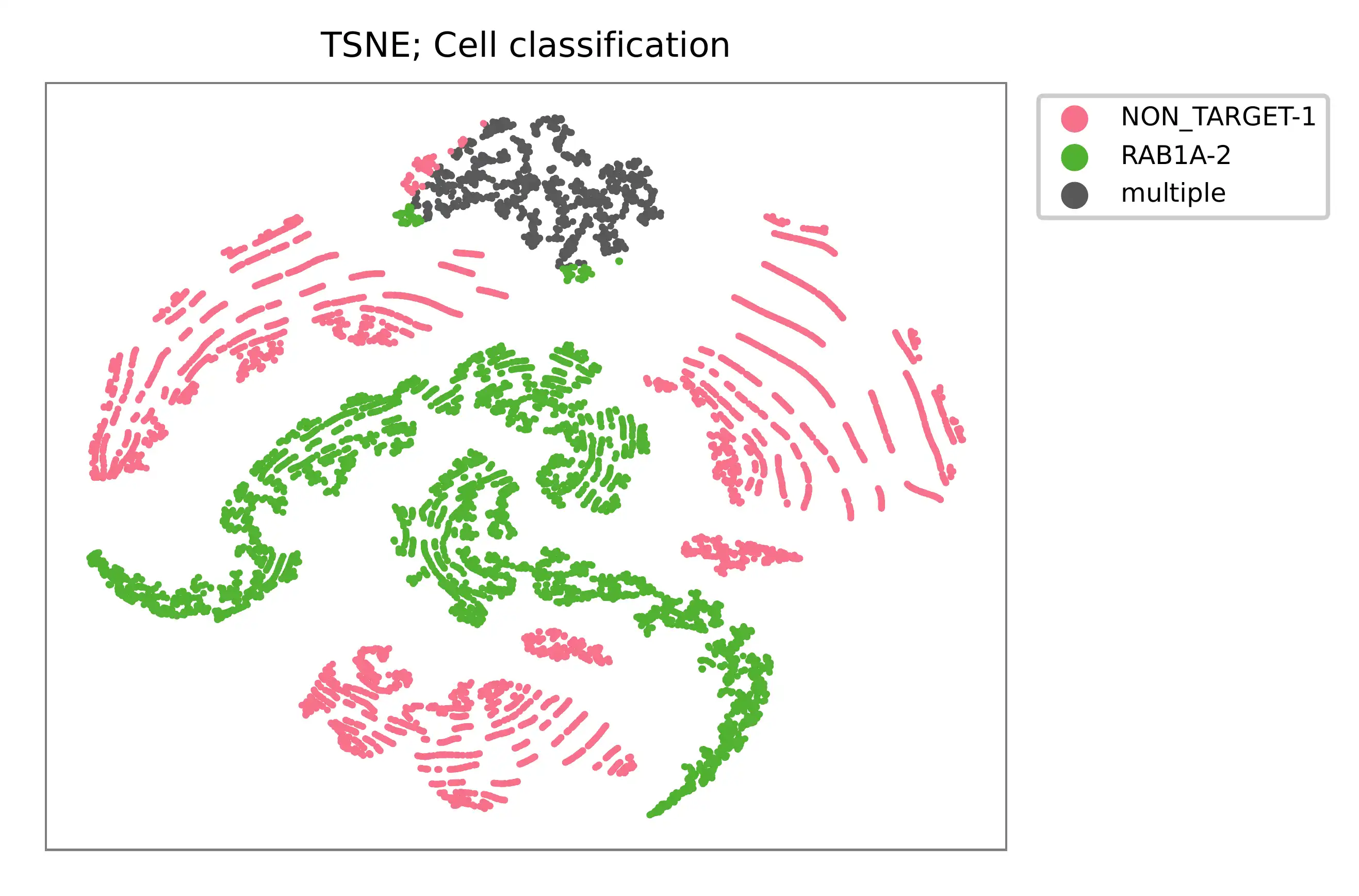
t-SNE embedding of cells based on the abundance of features (sgRNAs, no transcriptome information used). Colors indicate the sgRNA status for each cell, as called by FBA.
Gaussian mixture model¶
The implementation of demultiplexing method 2 (set by -dm) is
inspired by the method described on the 10x Genomics’ website. To set
the probability threshold for demultiplexing, use -p (default
0.9).
$ fba demultiplex \
-i matrix_featurecount.csv.gz \
-dm 2 \
-v
2021-10-04 14:14:15,659 - fba.__main__ - INFO - fba version: 0.0.x
2021-10-04 14:14:15,659 - fba.__main__ - INFO - Initiating logging ...
2021-10-04 14:14:15,659 - fba.__main__ - INFO - Python version: 3.8
2021-10-04 14:14:15,659 - fba.__main__ - INFO - Using demultiplex subcommand ...
2021-10-04 14:14:36,166 - fba.__main__ - INFO - Skipping arguments: "-q/--quantile", "-cm/--clustering_method"
2021-10-04 14:14:36,166 - fba.demultiplex - INFO - Output directory: demultiplexed
2021-10-04 14:14:36,166 - fba.demultiplex - INFO - Demultiplexing method: 2
2021-10-04 14:14:36,166 - fba.demultiplex - INFO - UMI normalization method: clr
2021-10-04 14:14:36,167 - fba.demultiplex - INFO - Visualization: On
2021-10-04 14:14:36,167 - fba.demultiplex - INFO - Visualization method: tsne
2021-10-04 14:14:36,167 - fba.demultiplex - INFO - Loading feature count matrix: matrix_featurecount.csv.gz ...
2021-10-04 14:14:37,875 - fba.demultiplex - INFO - Number of cells: 11,758
2021-10-04 14:14:37,875 - fba.demultiplex - INFO - Number of positive cells for a feature to be included: 200
2021-10-04 14:14:37,920 - fba.demultiplex - INFO - Number of features: 2 / 2 (after filtering / original in the matrix)
2021-10-04 14:14:37,920 - fba.demultiplex - INFO - Features: NON_TARGET-1 RAB1A-2
2021-10-04 14:14:37,920 - fba.demultiplex - INFO - Total UMIs: 7,145,799 / 7,145,799
2021-10-04 14:14:37,942 - fba.demultiplex - INFO - Median number of UMIs per cell: 477.0 / 477.0
2021-10-04 14:14:37,942 - fba.demultiplex - INFO - Demultiplexing ...
2021-10-04 14:14:38,418 - fba.demultiplex - INFO - Generating heatmap ...
2021-10-04 14:14:42,078 - fba.demultiplex - INFO - Embedding ...
2021-10-04 14:15:24,288 - fba.__main__ - INFO - Done.
Heatmap of the relative abundance of features (sgRNAs) across all cells. Each column represents a single cell.
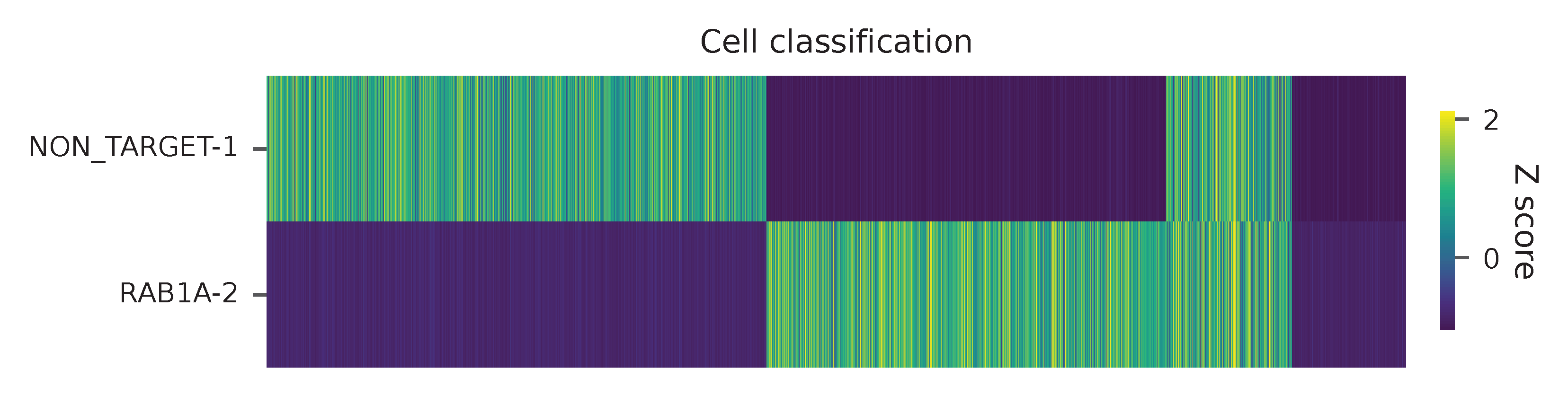
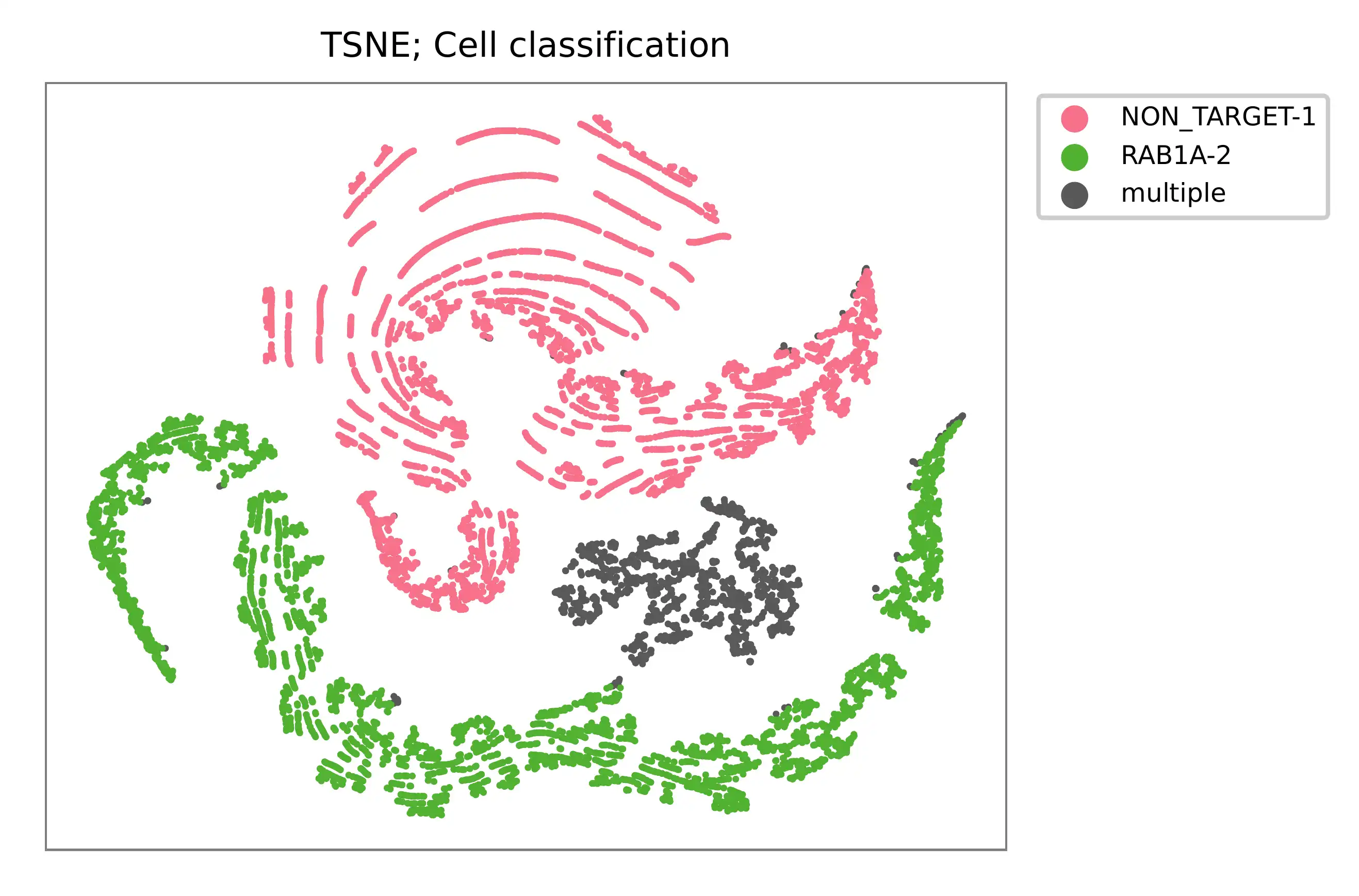
t-SNE embedding of cells based on the abundance of features (sgRNAs, no transcriptome information used). Colors indicate the sgRNA status for each cell, as called by FBA.
UMI distribution and model fitting threshold:
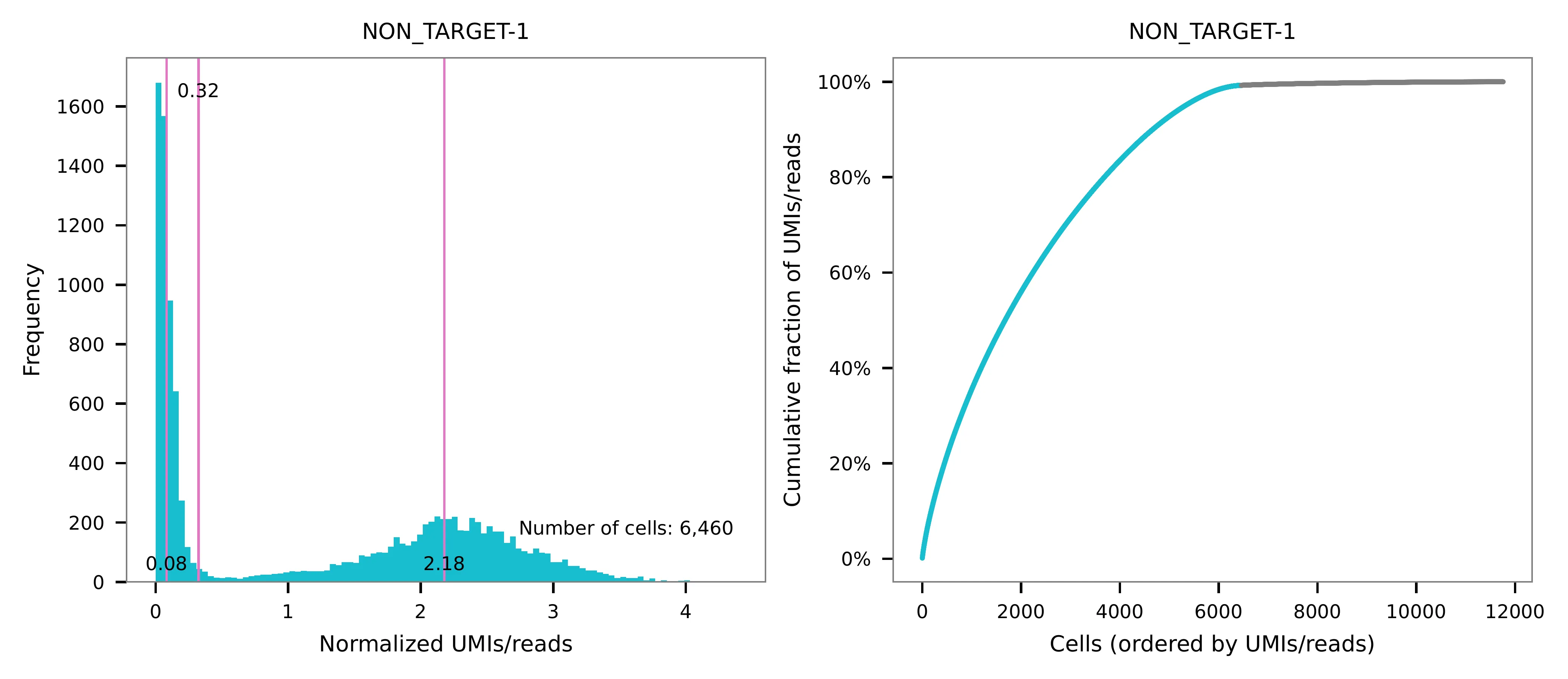
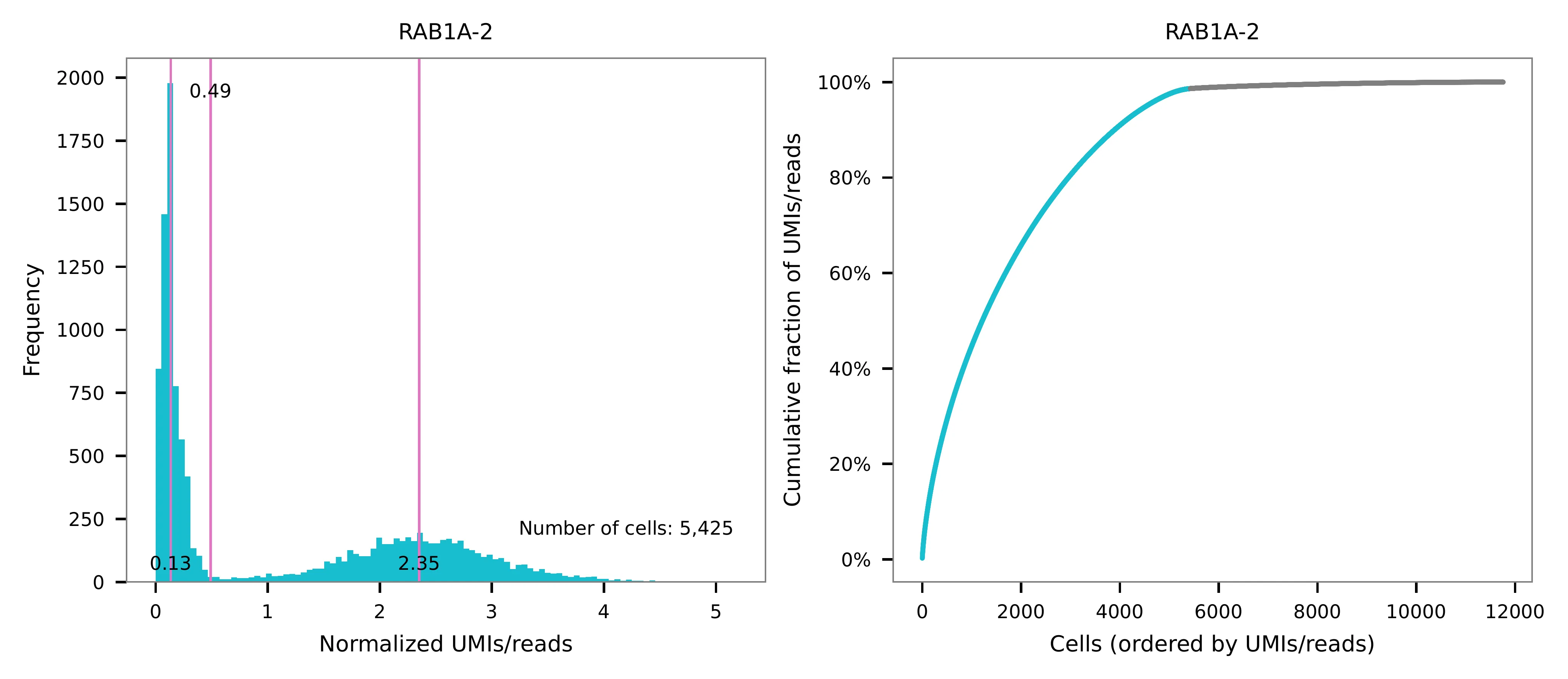
Poisson-Gaussian mixture model¶
The implementation of demultiplexing method 3 (set by -dm) is
inspired by Replogle, M., et al. (2021). The probability threshold
for demultiplexing can be set using -p (default 0.5).
$ fba demultiplex \
-i matrix_featurecount.csv.gz \
-dm 3 \
-v
2021-12-20 00:13:17,443 - fba.__main__ - INFO - fba version: 0.0.x
2021-12-20 00:13:17,443 - fba.__main__ - INFO - Initiating logging ...
2021-12-20 00:13:17,443 - fba.__main__ - INFO - Python version: 3.9
2021-12-20 00:13:17,443 - fba.__main__ - INFO - Using demultiplex subcommand ...
2021-12-20 00:13:19,774 - fba.__main__ - INFO - Skipping arguments: "-q/--quantile", "-cm/--clustering_method"
2021-12-20 00:13:19,774 - fba.demultiplex - INFO - Output directory: demultiplexed
2021-12-20 00:13:19,774 - fba.demultiplex - INFO - Demultiplexing method: 3
2021-12-20 00:13:19,774 - fba.demultiplex - INFO - UMI normalization method: clr
2021-12-20 00:13:19,774 - fba.demultiplex - INFO - Visualization: On
2021-12-20 00:13:19,774 - fba.demultiplex - INFO - Visualization method: tsne
2021-12-20 00:13:19,774 - fba.demultiplex - INFO - Loading feature count matrix: matrix_featurecount.csv.gz ...
2021-12-20 00:13:20,479 - fba.demultiplex - INFO - Number of cells: 11,758
2021-12-20 00:13:20,479 - fba.demultiplex - INFO - Number of positive cells for a feature to be included: 200
2021-12-20 00:13:20,497 - fba.demultiplex - INFO - Number of features: 2 / 2 (after filtering / original in the matrix)
2021-12-20 00:13:20,497 - fba.demultiplex - INFO - Features: NON_TARGET-1 RAB1A-2
2021-12-20 00:13:20,497 - fba.demultiplex - INFO - Total UMIs: 7,145,799 / 7,145,799
2021-12-20 00:13:20,506 - fba.demultiplex - INFO - Median number of UMIs per cell: 477.0 / 477.0
2021-12-20 00:13:20,506 - fba.demultiplex - INFO - Demultiplexing ...
2021-12-20 00:13:21,930 - fba.demultiplex - INFO - Generating heatmap ...
2021-12-20 00:13:23,070 - fba.demultiplex - INFO - Embedding ...
2021-12-20 00:13:41,271 - fba.__main__ - INFO - Done.
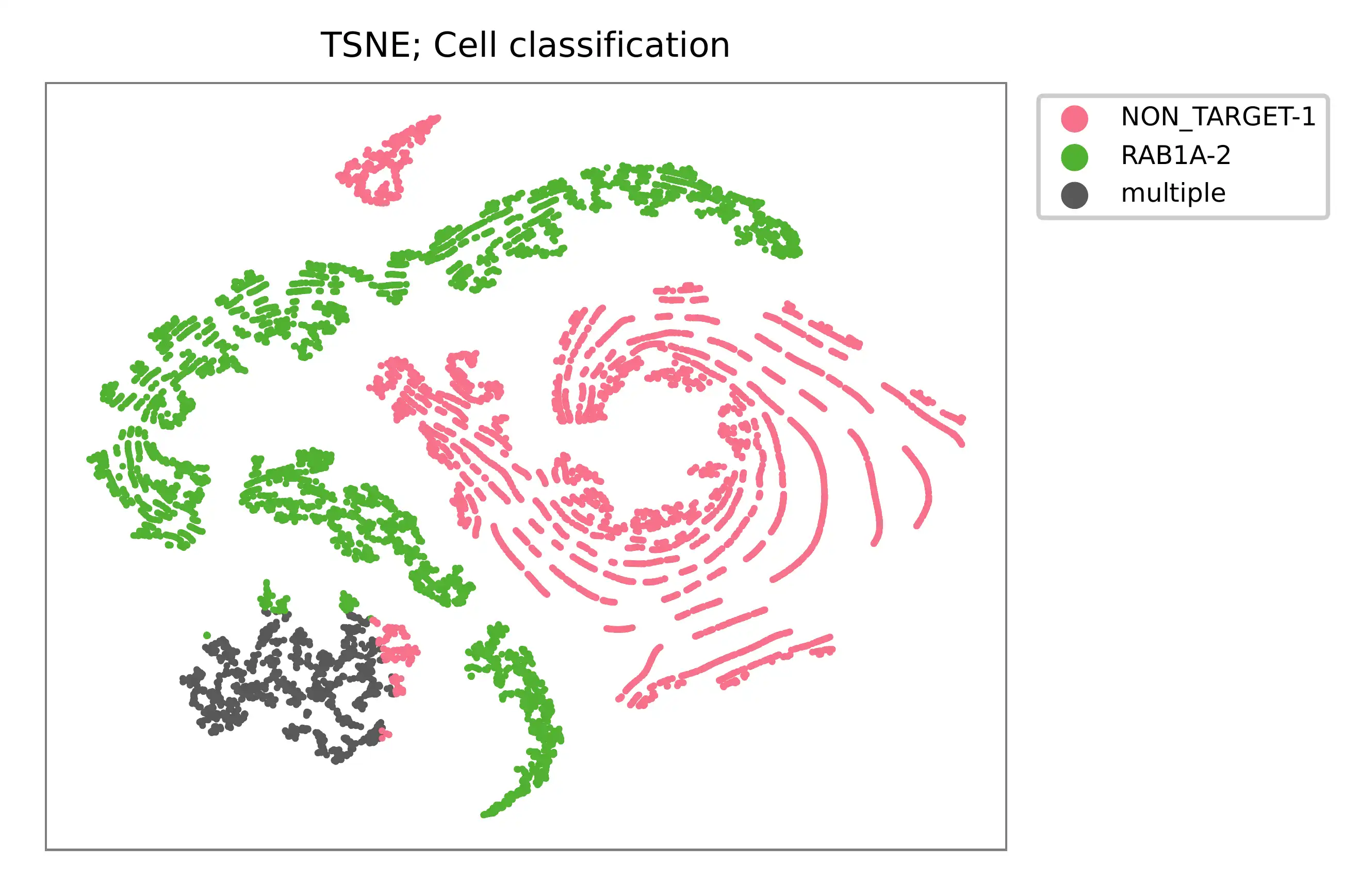
Heatmap of the relative abundance of features (sgRNAs) across all cells. Each column represents a single cell.
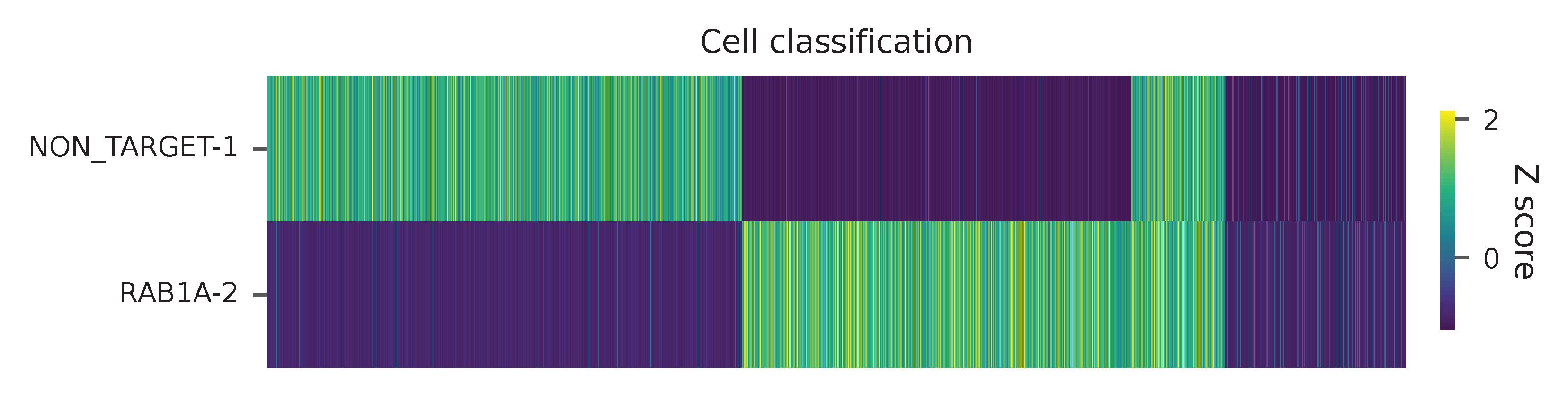
t-SNE embedding of cells based on the abundance of features (sgRNAs, no transcriptome information used). Colors indicate the sgRNA status for each cell, as called by FBA.
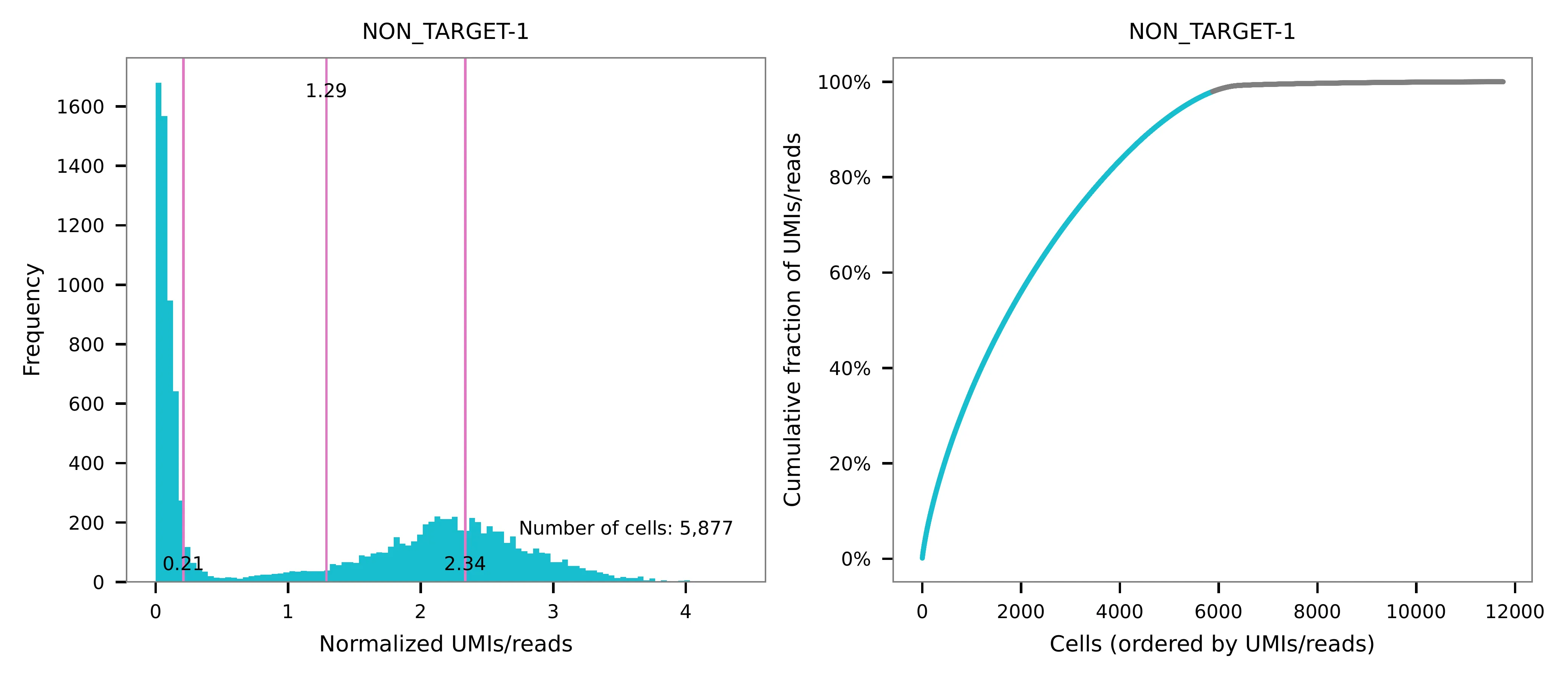
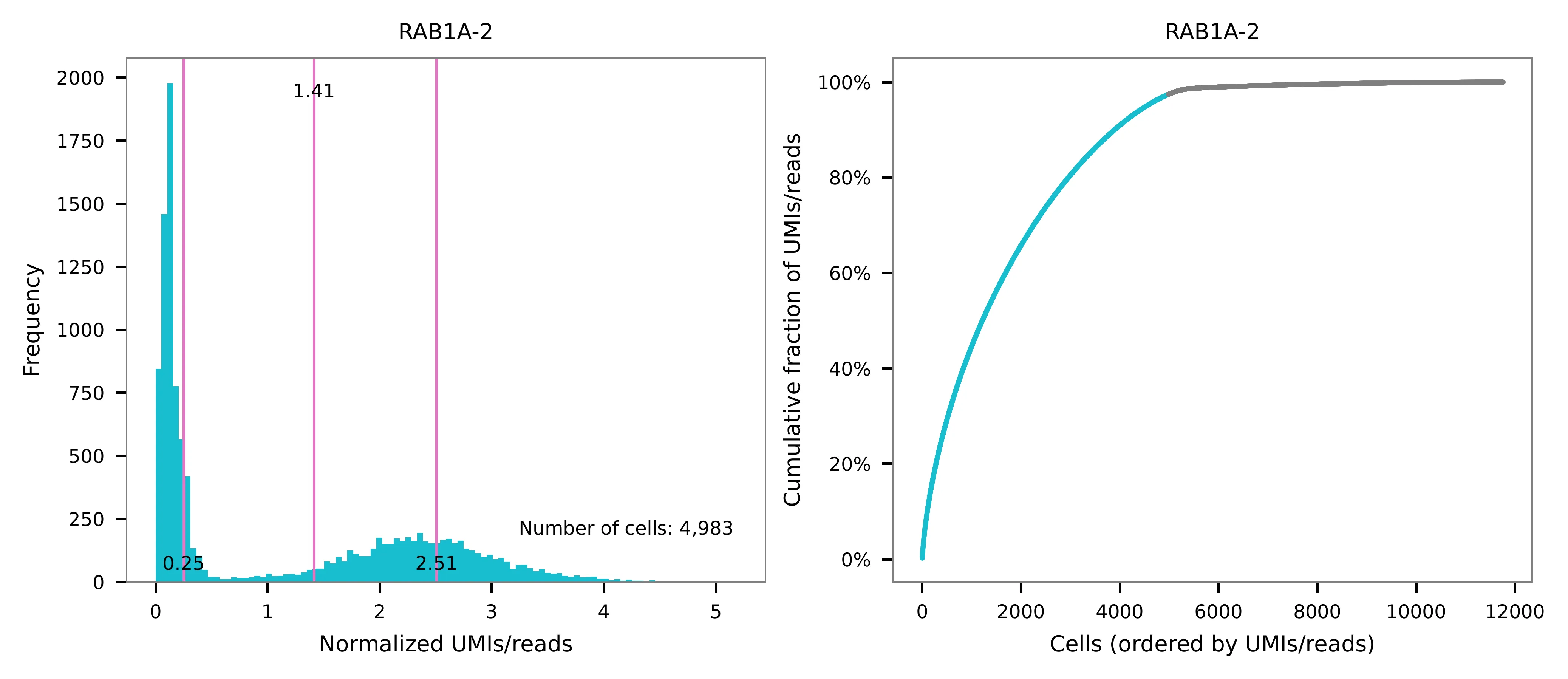
UMI distribution and model fitting threshold:
Kernel density estimation¶
CRISPR perturbations are demultiplexed based on the abundance of
features using demultiplexing method 4, which is implemented with
modifications to the method described in McGinnis, C., et al. (2019).
$ fba demultiplex \
-i matrix_featurecount.csv.gz \
-dm 4 \
-v
2021-12-26 18:11:16,685 - fba.__main__ - INFO - fba version: 0.0.x
2021-12-26 18:11:16,685 - fba.__main__ - INFO - Initiating logging ...
2021-12-26 18:11:16,686 - fba.__main__ - INFO - Python version: 3.9
2021-12-26 18:11:16,686 - fba.__main__ - INFO - Using demultiplex subcommand ...
2021-12-26 18:11:19,633 - fba.__main__ - INFO - Skipping arguments: "-q/--quantile", "-cm/--clustering_method", "-p/--prob"
2021-12-26 18:11:19,633 - fba.demultiplex - INFO - Output directory: demultiplexed
2021-12-26 18:11:19,633 - fba.demultiplex - INFO - Demultiplexing method: 4
2021-12-26 18:11:19,633 - fba.demultiplex - INFO - UMI normalization method: clr
2021-12-26 18:11:19,633 - fba.demultiplex - INFO - Visualization: On
2021-12-26 18:11:19,633 - fba.demultiplex - INFO - Visualization method: tsne
2021-12-26 18:11:19,633 - fba.demultiplex - INFO - Loading feature count matrix: matrix_featurecount.csv.gz ...
2021-12-26 18:11:19,745 - fba.demultiplex - INFO - Number of cells: 11,758
2021-12-26 18:11:19,745 - fba.demultiplex - INFO - Number of positive cells for a feature to be included: 200
2021-12-26 18:11:19,762 - fba.demultiplex - INFO - Number of features: 2 / 2 (after filtering / original in the matrix)
2021-12-26 18:11:19,762 - fba.demultiplex - INFO - Features: NON_TARGET-1 RAB1A-2
2021-12-26 18:11:19,762 - fba.demultiplex - INFO - Total UMIs: 7,145,799 / 7,145,799
2021-12-26 18:11:19,771 - fba.demultiplex - INFO - Median number of UMIs per cell: 477.0 / 477.0
2021-12-26 18:11:19,771 - fba.demultiplex - INFO - Demultiplexing ...
2021-12-26 18:11:22,049 - fba.demultiplex - INFO - Quantile cutoff: 18
2021-12-26 18:11:23,703 - fba.demultiplex - INFO - Generating heatmap ...
2021-12-26 18:11:24,911 - fba.demultiplex - INFO - Embedding ...
2021-12-26 18:11:44,219 - fba.__main__ - INFO - Done.
Heatmap of the relative abundance of features (sgRNAs) across all cells. Each column represents a single cell.
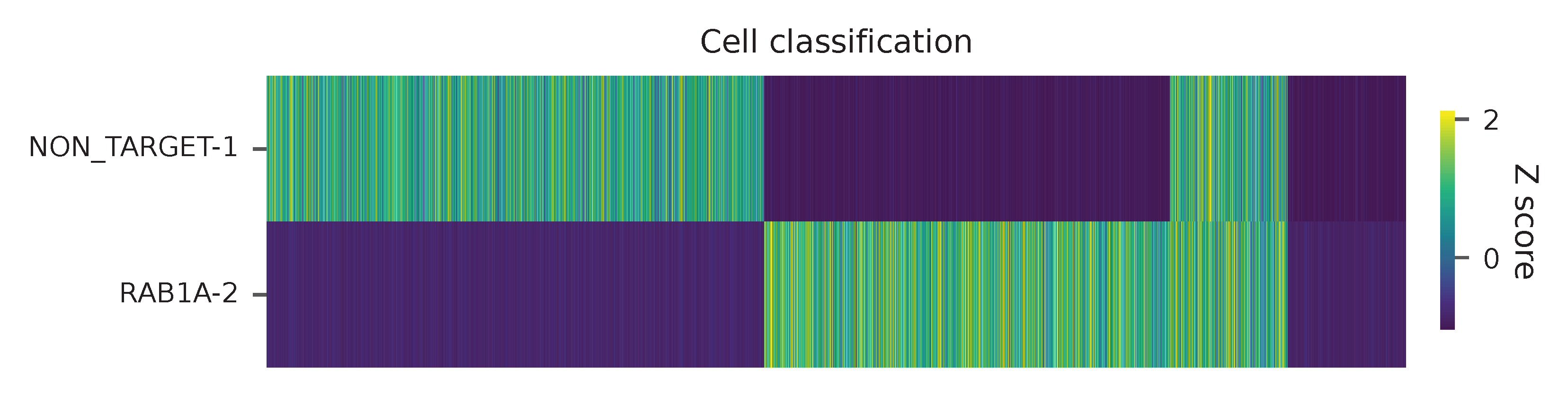
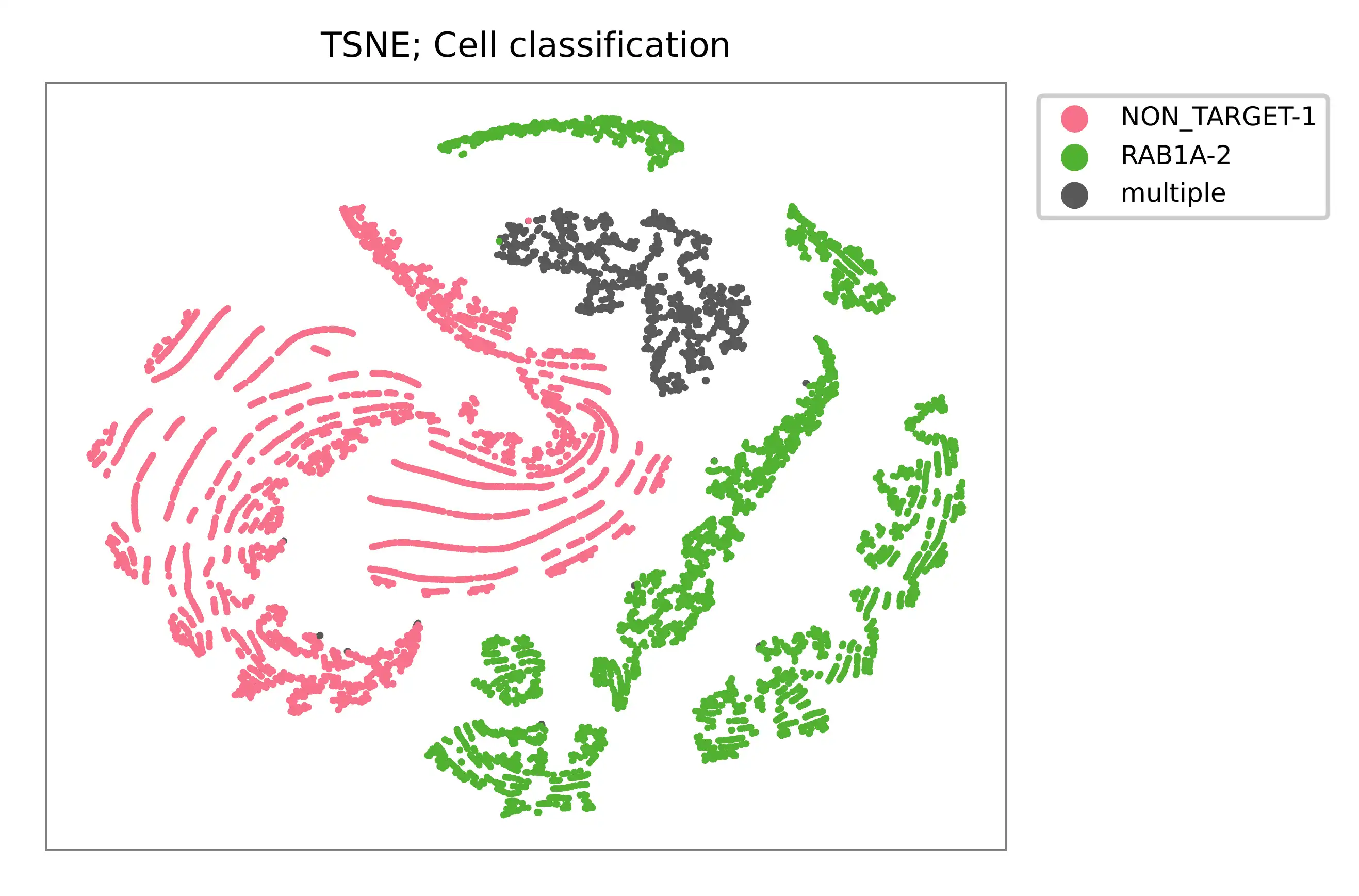
t-SNE embedding of cells based on the abundance of features (sgRNAs, no transcriptome information used). Colors indicate the sgRNA status for each cell, as called by FBA.
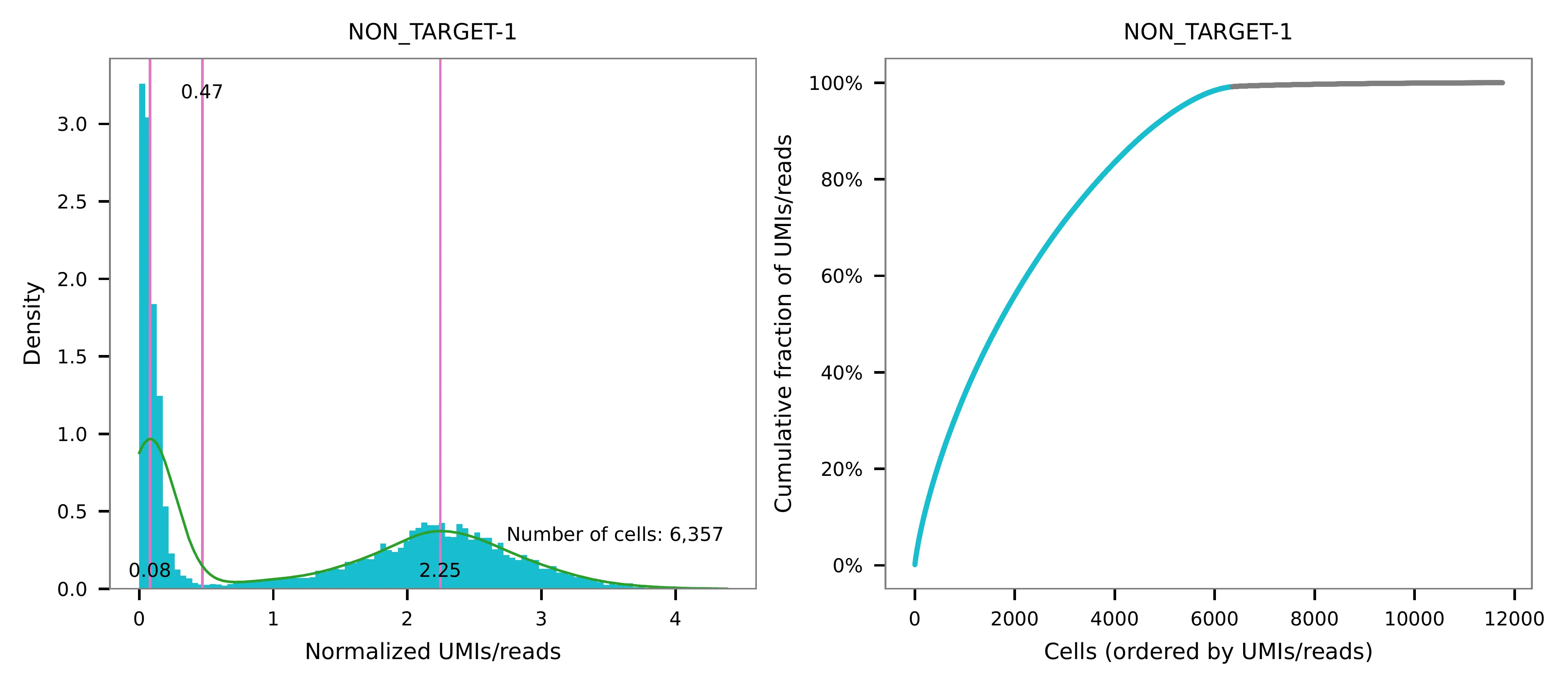
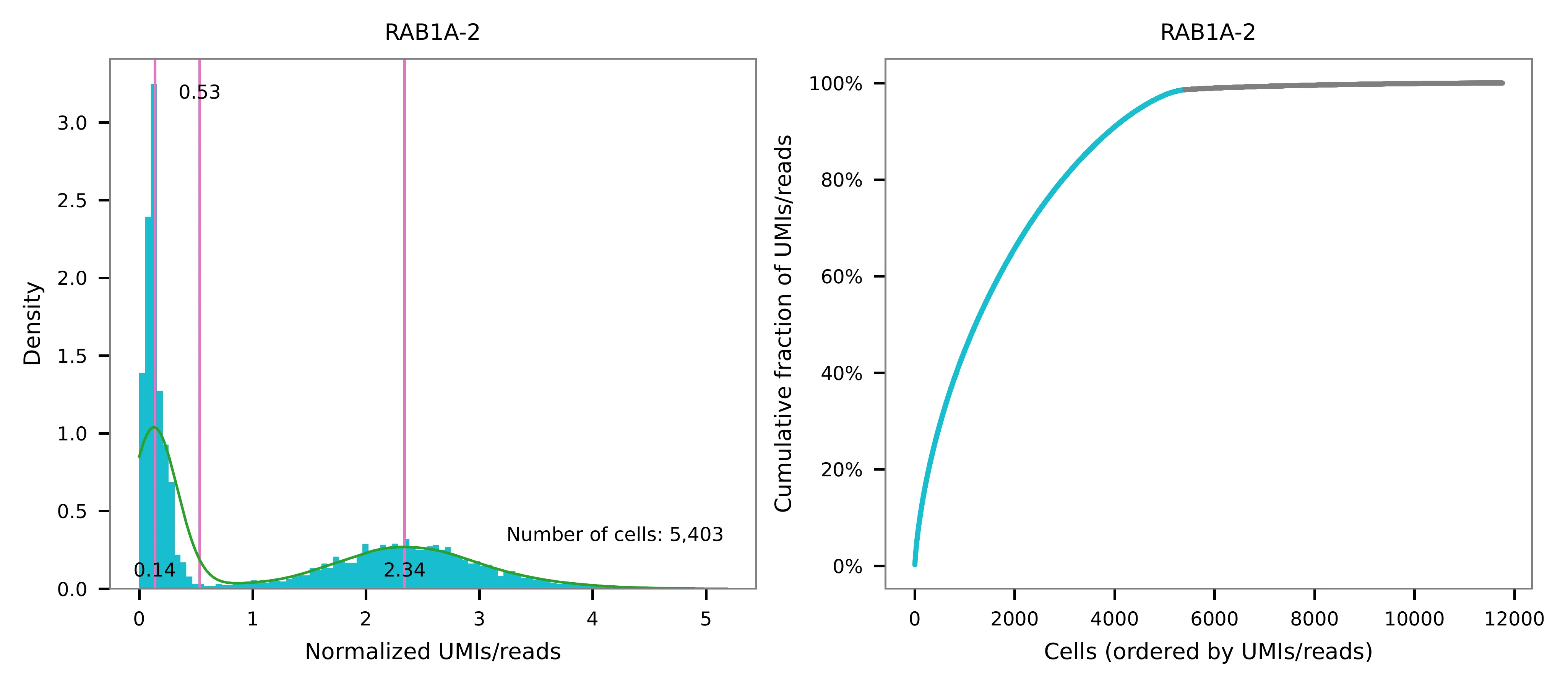
UMI distribution and model fitting threshold:
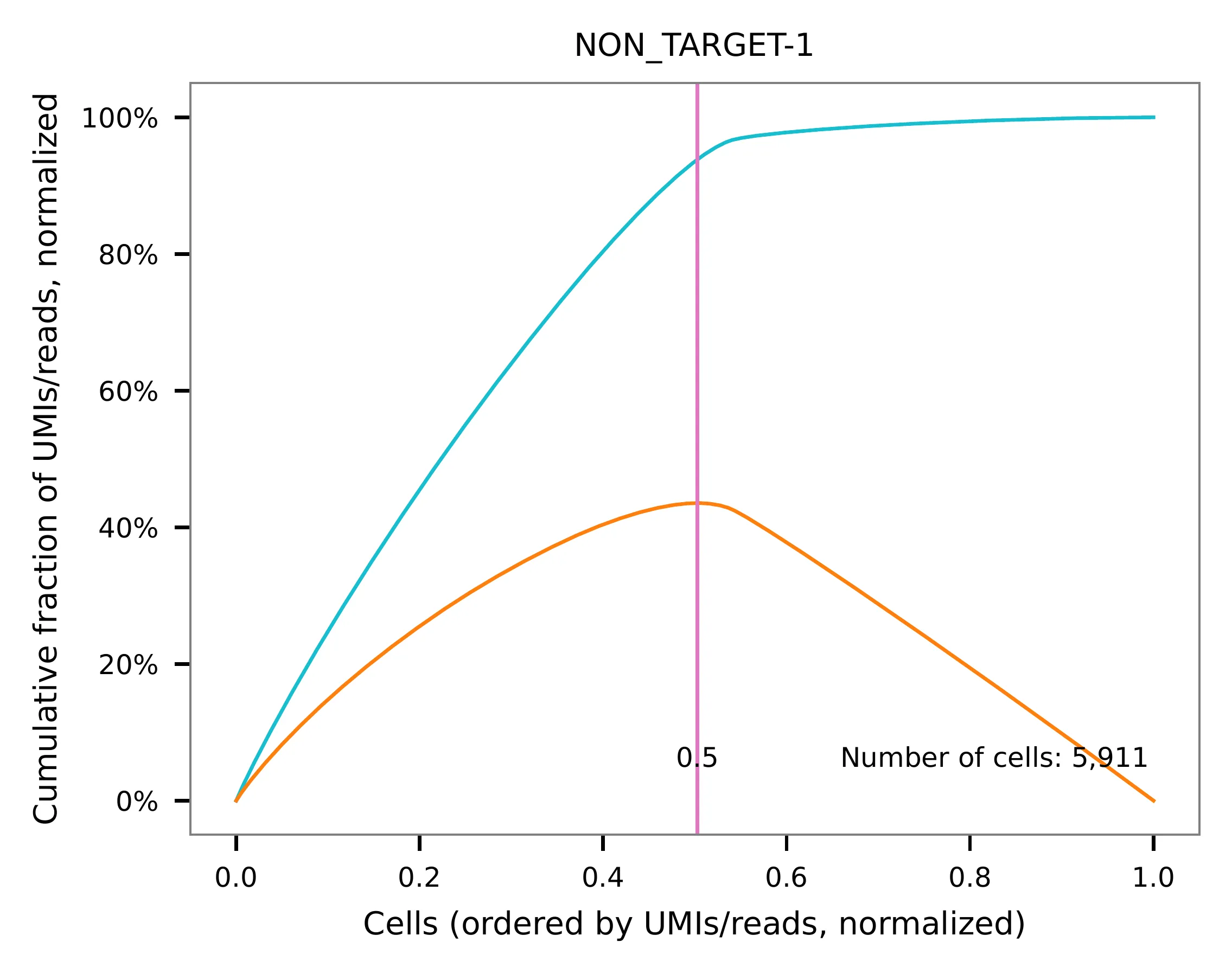
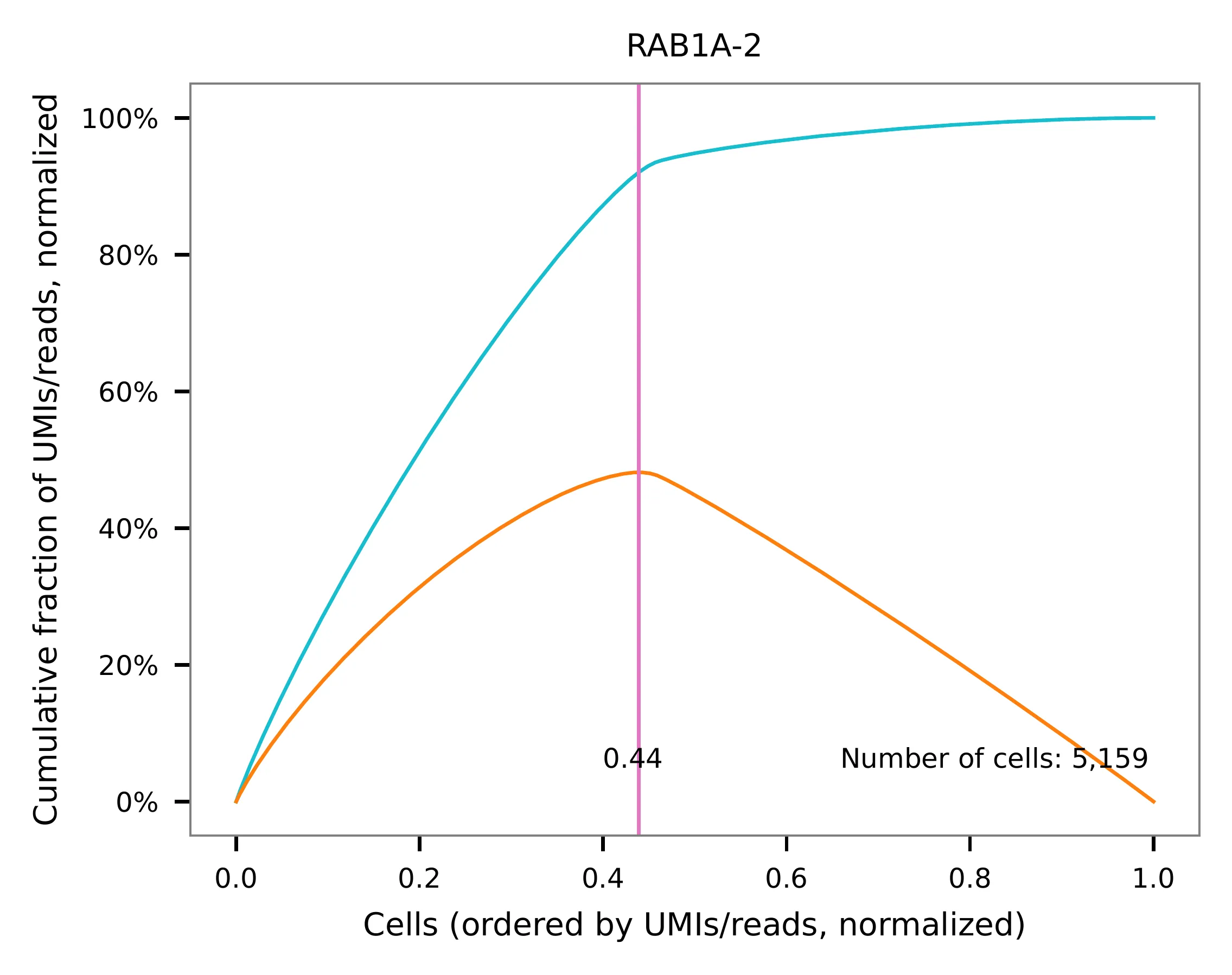
Knee point¶
Method 1¶
Cells are demultiplexed based on the abundance of features, specifically
sgRNAs. Demultiplexing method 5-2019 is our previous implementation,
which aims to identify perturbations in the cells by detecting an
inflection point on the feature UMI saturation curve (Xie, S., et al.
2019).
$ fba demultiplex \
-i matrix_featurecount.csv.gz \
-dm 5-2019 \
-v
2022-01-02 13:57:51,792 - fba.__main__ - INFO - fba version: 0.0.x
2022-01-02 13:57:51,792 - fba.__main__ - INFO - Initiating logging ...
2022-01-02 13:57:51,792 - fba.__main__ - INFO - Python version: 3.9
2022-01-02 13:57:51,792 - fba.__main__ - INFO - Using demultiplex subcommand ...
2022-01-02 13:57:54,328 - fba.__main__ - INFO - Skipping arguments: "-q/--quantile", "-cm/--clustering_method", "-p/--prob"
2022-01-02 13:57:54,329 - fba.demultiplex - INFO - Output directory: demultiplexed
2022-01-02 13:57:54,329 - fba.demultiplex - INFO - Demultiplexing method: 5-2019
2022-01-02 13:57:54,329 - fba.demultiplex - INFO - UMI normalization method: clr
2022-01-02 13:57:54,329 - fba.demultiplex - INFO - Visualization: On
2022-01-02 13:57:54,329 - fba.demultiplex - INFO - Visualization method: tsne
2022-01-02 13:57:54,329 - fba.demultiplex - INFO - Loading feature count matrix: raw/m2_2020-10-20/matrix_featurecount.csv.gz ...
2022-01-02 13:57:54,444 - fba.demultiplex - INFO - Number of cells: 11,758
2022-01-02 13:57:54,444 - fba.demultiplex - INFO - Number of positive cells for a feature to be included: 200
2022-01-02 13:57:54,464 - fba.demultiplex - INFO - Number of features: 2 / 2 (after filtering / original in the matrix)
2022-01-02 13:57:54,464 - fba.demultiplex - INFO - Features: NON_TARGET-1 RAB1A-2
2022-01-02 13:57:54,464 - fba.demultiplex - INFO - Total UMIs: 7,145,799 / 7,145,799
2022-01-02 13:57:54,474 - fba.demultiplex - INFO - Median number of UMIs per cell: 477.0 / 477.0
2022-01-02 13:57:54,474 - fba.demultiplex - INFO - Demultiplexing ...
2022-01-02 13:57:55,509 - fba.demultiplex - INFO - Generating heatmap ...
2022-01-02 13:57:56,717 - fba.demultiplex - INFO - Embedding ...
2022-01-02 13:58:12,014 - fba.__main__ - INFO - Done.
Heatmap of the relative abundance of features (sgRNAs) across all cells. Each column represents a single cell.
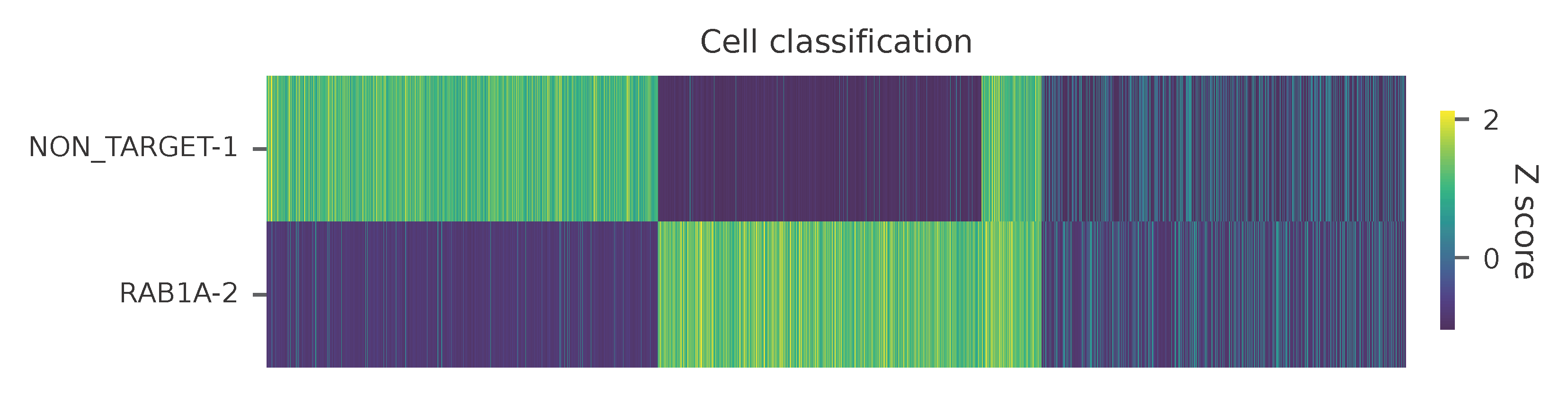
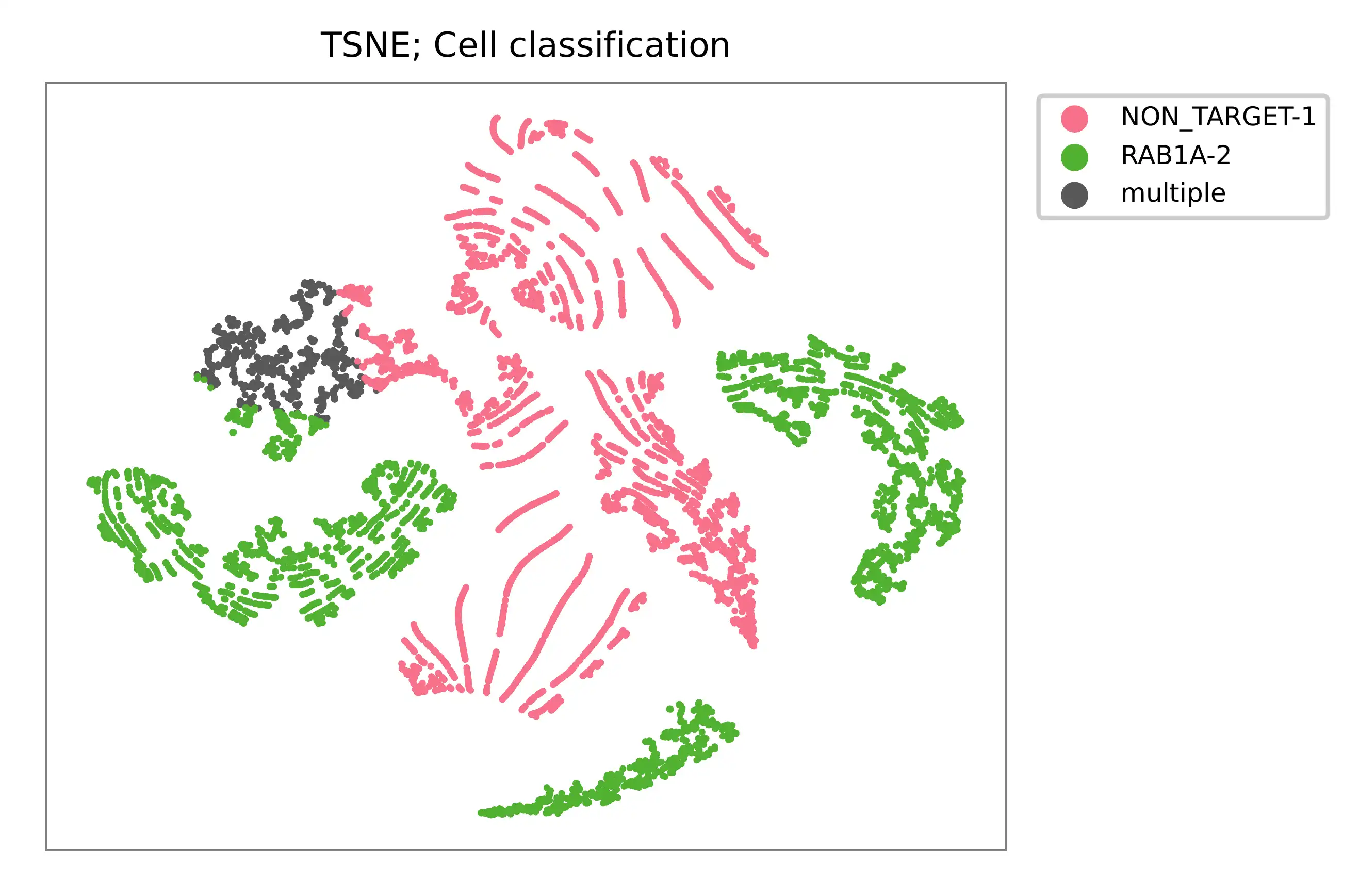
t-SNE embedding of cells based on the abundance of features (sgRNAs, no transcriptome information used). Colors indicate the sgRNA status for each cell, as called by FBA.
UMI distribution and model fitting threshold:
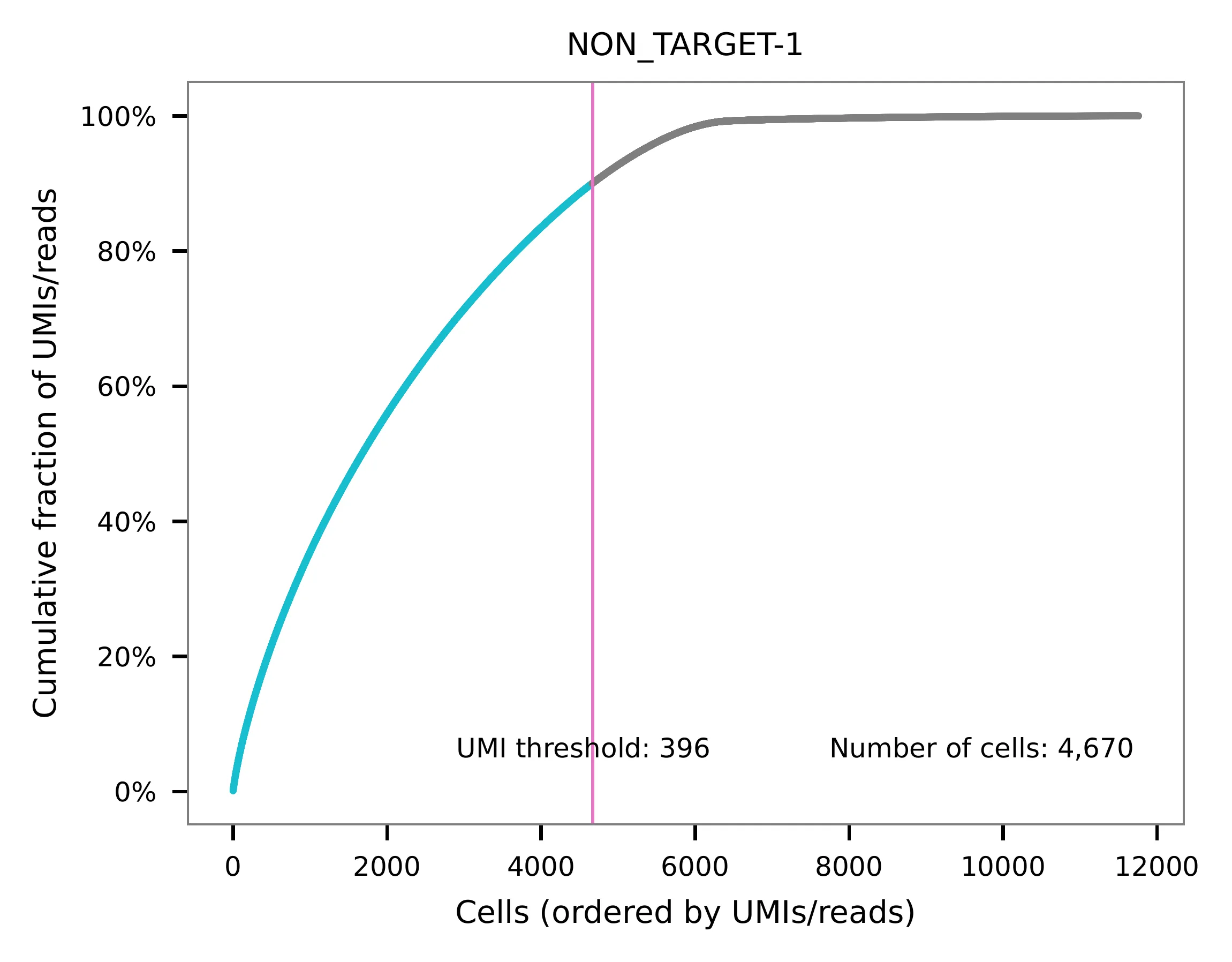
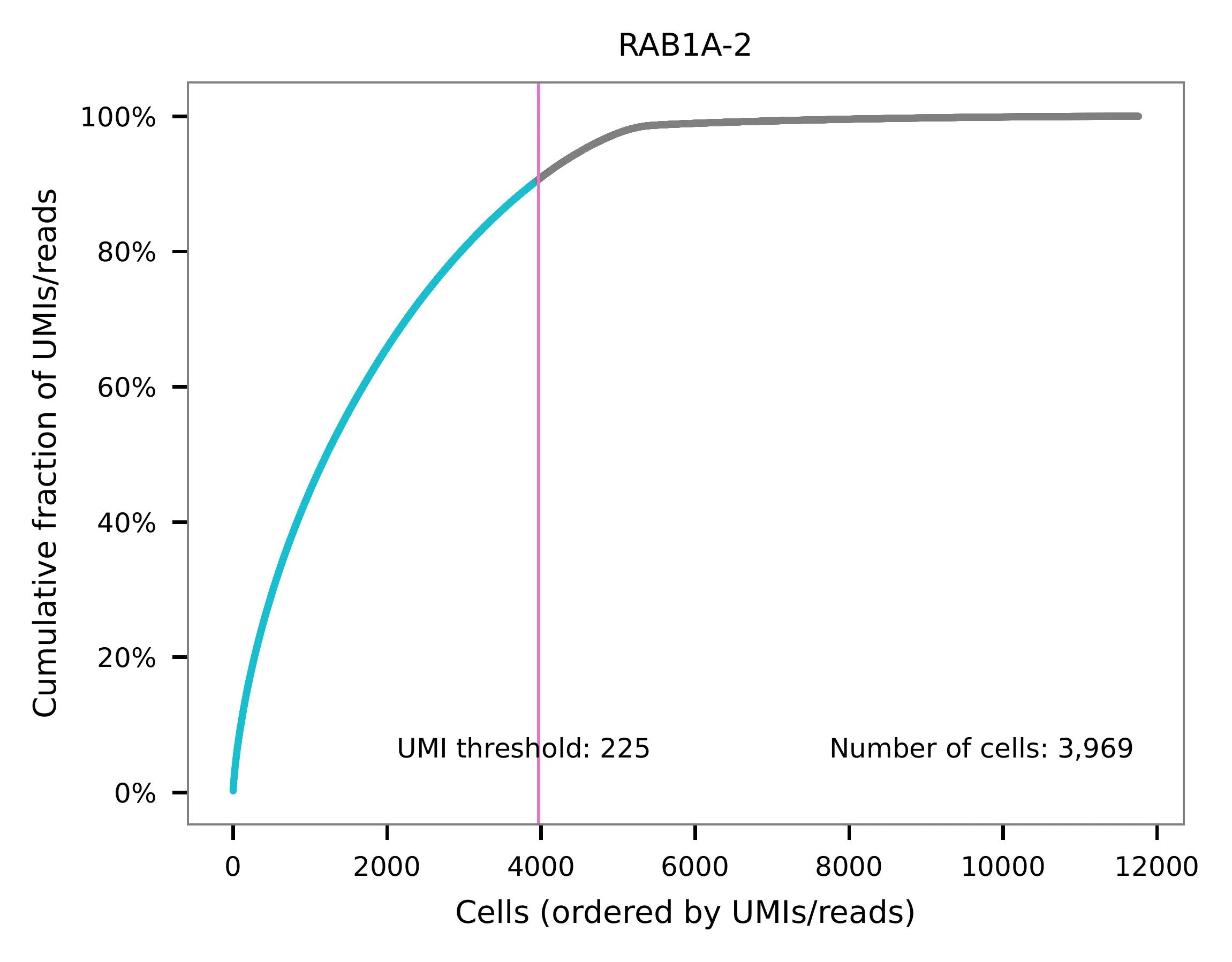
Method 2¶
Cells are demultiplexed based on the abundance of features, specifically
sgRNAs. Demultiplexing method 5 is implemented to use the local
maxima on the difference curve to detemine the knee point on the UMI
saturation curve.
$ fba demultiplex \
-i matrix_featurecount.csv.gz \
-dm 5 \
-v
2021-12-29 22:55:34,303 - fba.__main__ - INFO - fba version: 0.0.x
2021-12-29 22:55:34,303 - fba.__main__ - INFO - Initiating logging ...
2021-12-29 22:55:34,303 - fba.__main__ - INFO - Python version: 3.9
2021-12-29 22:55:34,303 - fba.__main__ - INFO - Using demultiplex subcommand ...
2021-12-29 22:55:36,774 - fba.__main__ - INFO - Skipping arguments: "-q/--quantile", "-cm/--clustering_method", "-p/--prob"
2021-12-29 22:55:36,774 - fba.demultiplex - INFO - Output directory: demultiplexed
2021-12-29 22:55:36,774 - fba.demultiplex - INFO - Demultiplexing method: 5
2021-12-29 22:55:36,774 - fba.demultiplex - INFO - UMI normalization method: clr
2021-12-29 22:55:36,774 - fba.demultiplex - INFO - Visualization: On
2021-12-29 22:55:36,774 - fba.demultiplex - INFO - Visualization method: tsne
2021-12-29 22:55:36,774 - fba.demultiplex - INFO - Loading feature count matrix: matrix_featurecount.csv.gz ...
2021-12-29 22:55:36,886 - fba.demultiplex - INFO - Number of cells: 11,758
2021-12-29 22:55:36,886 - fba.demultiplex - INFO - Number of positive cells for a feature to be included: 200
2021-12-29 22:55:36,904 - fba.demultiplex - INFO - Number of features: 2 / 2 (after filtering / original in the matrix)
2021-12-29 22:55:36,904 - fba.demultiplex - INFO - Features: NON_TARGET-1 RAB1A-2
2021-12-29 22:55:36,904 - fba.demultiplex - INFO - Total UMIs: 7,145,799 / 7,145,799
2021-12-29 22:55:36,913 - fba.demultiplex - INFO - Median number of UMIs per cell: 477.0 / 477.0
2021-12-29 22:55:36,913 - fba.demultiplex - INFO - Demultiplexing ...
2021-12-29 22:55:37,415 - fba.demultiplex - INFO - Generating heatmap ...
2021-12-29 22:55:38,576 - fba.demultiplex - INFO - Embedding ...
2021-12-29 22:55:57,485 - fba.__main__ - INFO - Done.
Heatmap of the relative abundance of features (sgRNAs) across all cells. Each column represents a single cell.
t-SNE embedding of cells based on the abundance of features (sgRNAs, no transcriptome information used). Colors indicate the sgRNA status for each cell, as called by FBA.
UMI distribution and model fitting threshold: